True or false: if we want to achieve any degree of semantic interoperability in our clinical systems we need to standardise the clinical content, keeping it open and independent of any single implementation or messaging formalism?
Therapeutic Precautions are the 'new black'!
Clinical modelling around the concepts of warnings, alerts and notifications is incredibly complex and each of the terms are loaded with confusion. It is not going to be easy to navigate this area and achieve a common understanding that will underpin information models for sensible and cross paradigm decision support.
The Archetype Journey...
I'm surprised to realise I've been building archetypes for over 7 years. It honestly doesn't feel that long. It still feels like we are in the relatively early days of understanding how to model clinical archetypes, to validate them and to govern them. I am learning more with each archetype I build. They are definitely getting better and the process more refined. But we aren't there yet. We have a ways to go! Let me try to share some idea of the challenges and complexities I see…
We can build all kinds of archetypes for different purposes.
There are the ones we just want to use for our own project or purpose, to be used in splendid isolation. Yes, anyone can build an archetype for any reason. Easy as. No design constraints, no collaboration, just whatever you want to model and as large or complex as you like.
But if you want to build them so that they will be re-used and shared, then a whole different approach is required. Each archetype needs to fit with the others around it, to complement but not duplicate or overlap; to be of the same granularity; to be consistent with the way similar concepts are modelled; to have the same principles regarding the level of detail modelled; the same approach to defining scope; and of course the same approach to defining a clinical concept versus a data element or group of data elements… The list goes on.
Some archetypes are straightforward to design and build, for example all the very prescriptive and well recognised scales like the Braden Scale or Glasgow Coma Scale. These are the 'no brainers' of clinical modelling.
Some are harder and more abstract, such as those underpinning a clinical decision support system of orders and activities to ensure that care plans are carried out, clinical outcomes achieved and patients don't 'fall through the cracks' from transitions of care.
Then there are the repositories of archetypes that are intended to work as single, cohesive pool of models – each archetype for a single clinical concept that all sits closely aligned to the next one, but minimising any duplication or overlap.
That is a massive coordination task, and one that I underestimated hugely when we embarked on the development of the openEHR Clinical Knowledge Manager, and especially more recently, the really active development and coordination required to manage the model development, collaboration and management process within the Australian CKM – where the national eHealth program and jurisdictions are working within the same domain of models, developing new ones for specific purposes and re-using common, shared models for different use cases and clinical contexts.
The archetype ecosystems are hard, numbers of archetypes that need to work together intimately and precisely to enable the accurate and safe modelling of clinical data. Physical examination is the perfect example that has been weighing on my mind now for some time. I've dabbled with small parts of this over the years, as specific projects needed to model a small part of the physical exam here and there. My initial focus was on modelling generic patterns for inspection, palpation, auscultation and percussion – four well identified pillars of the art of clinical examination. If you take a look at the Inspection archetype clinicians will recognise the kind of pattern that we were taught in First Year of our Medical or Nursing degrees. And I built huge mind maps to try to anticipate how the basic generic pattern could be specialised or adapted for use in all aspects of recording the inspection component of clinical examination. Over time, I have convinced myself that this would not work, and so the ongoing dilemma was how to approach it to create a standardised, yet extraordinarily flexible solution.
Over time, I have convinced myself that this would not work, and so the ongoing dilemma was how to approach it to create a standardised, yet extraordinarily flexible solution.
Consider the dilemma of modelling physical examination. How can we capture the fractal nature of physical examination? How can we represent the art of every clinician's practice in standardised archetypes? We need models that can be standardised, yet we also need to be able to respond to the massive variability in the requirements and approach of each and every clinician. Each profession will record the same concept in different levels of detail, and often in a slightly different context. Each specialty will record different combinations of details. Specialists need all the detail; generalists only want to record the bare basics, unless they find something significant in which case they want to drill down to the nth degree. And don't forget the ability to just quickly note 'NAD' as you fly past to the next part of the examination; for rheumatologists to record a homunculus; for the requirement for addition of photos or annotated diagrams! Ha – modelling physical examination IS NOT SIMPLE!
I think I might have finally broken the back of the physical examination modelling dilemma just this week. Seven years after starting this journey, with all this modelling experience behind me! The one sure thing I have learned – a realisation of how much we don't know. Don't let anyone tell you it is easy or we know enough. IMO we aren't ready to publish standards or even specifications about this work, yet. But we are making good, sound, robust progress. We can start to document our experience and sound principles.
This new domain of clinical knowledge management is complex; nobody should be saying we have it sorted...
The ultimate PHR?
I've been interested in the notion of a Personal Health Record for a long time. I was involved in the development of HotHealth, which launched at the end of 2000, a not-so-auspicious year, given the dot com crash! By the time HotHealth was completed , all the potential competitors identified in the pre-market environmental scan were defunct. It certainly wasn't easy to get any traction for HotHealth take-up and yet only recently it has been retired. For a couple of years it was successfully used at the Royal Children's Hospital, cut down and re-branded as BetterDiabetes to support teenagers self-manage their diabetes and communicate with their clinicians, but it wasn't sustained.
This is not an uncommon story for PHRs. It is somewhat comforting to see that the course of those such HealthVault and GoogleHealth have also not been smooth and fabulously successful :-)
Why is the PHR so hard?
In recent years I participated in the development of the ISO Technical Report 14292:2012: Personal health records -- Definition, scope and context. In this my major contribution seemed to be introducing the idea of a health information continuum.
However in the past year or so, my notion of an ideal PHR has moved on a little further again. It has arisen on the premise of a health record platform in which standardised health information persists independently of any one software application and can be accessed by any compliant applications, whether consumer- or clinician-focused. And the record of health information can be contributed to by any number of compliant systems - whether a clinical system, a PHR or smartphone app. The focus is on the data, the health record itself; not the applications. You will have seen a number of my previous posts, including here & here!
So, in this kind of new health data utopia, imagine if all my weights were automatically uploaded to my Weight app on my smartphone wirelessly each morning. Over time I could graph this and track my BMI etc. Useful stuff, and this can be done now - but only into dead-end silos of data within a given app.
And what if a new fandangled weight management application came along that I liked better - perhaps it provided more support to help me lose weight. And I want to lose weight. So I add the new app to my smartphone and, hey presto, it can immediately access all my previous weights - all because the data structure in both apps is identical. Thus the data can be unambiguously understood and computed upon within the second app without any data manipulation. Pretty cool. No more data silos; no more data loss. Simply delete the first app from the system, and elect to keep the data within my smartphone health record.
And as I add apps that suit my lifestyle, health needs, and fitness goals etc, I'm gradually accumulating important health information that is probably not available anywhere else. And consider that only I actually know what medicines I'm taking, including over the counter and herbals. The notion of a current medication list is really not in the remit of any clinician, but the motivated consumer! And so if I add an app to start to manage my medications or immunisations this data could be also used across in yet another compliant chronic disease support app for my diabetes or asthma or...
I can gradually build up a record of health information that is useful to me to manage my health, and that is also potentially useful to share with my healthcare providers.
Do you see the difference to current PHR systems?
I can choose apps that are 'best of breed' and applicable to my need or interest.
I'm not locked in to any one app, a mega app that contains stuff I don't want and will never use, with all the overheads and lack of flexibility.
I can 'plug & play' apps into my health record, able to change my mind if I find features, a user interface or workflow that I like better.
And yet the data remains ready for future use and potentially for sharing with my healthcare providers, if and when I choose. How cool is that?
Keep in mind that if those data structures were the same as being used by my clinician systems, then there is also potential for me to receive data from my clinicians and incorporate it into my PHR; similarly there is also potential for me to send data to my clinician and give them the choice of incorporating this into their systems - maybe my blood glucose records directly obtained from my glucometer, my weight measurements, etc. Maybe, one day, even MY current medicine list!
In this proposed flexible data environment we are avoiding the 'one size fits all', behemoth approach, which doesn't seem to have worked well in many situations, both clinical systems or personal health records. Best of all the data is preserved in the non-proprietary, shared format - the beginnings of a universal health record or, at least, a health record platform fully supporting data exchange.
What do you think?
Are we there yet?
No, but we are definitely moving in the right direction... Conversations are happening that were uncommon generally, and downright rare in the US only 18 months ago. I've been rabbiting on for some time about the need for a 'universal health record - an application-independent core of shared and standardised health information into which a variety of 'enlightened' applications can 'plug & play'; thus breaking down the hold of the proprietary and 'not invented here' approach of proprietary clinical applications with which we battle most everywhere today.
So it was pleasing to see Margalit Gur-Arie's recent blog post on Arguments for a Universal Health Record. While I'm not convinced about the reality a single database (see my comments at the end of Margalit's post), I wholeheartedly endorse the principle of having a single approach to defining the data - this is a very powerful concept, and one that may well become a pivotal enabler to health IT innovation.
In addition, Kevin Coonan has started blogging in recent days - see his Summary of DCMs regarding principles of Detailed Clinical Models (aka DCMs). Now I know that Kevin's vision for an implementable HL7 DCM is totally different to the openEHR DCMs (=archetypes) that I work with. But we do agree on the basic principles about the basic attributes of these models that he has outlined in his blog post - it is quite a good summary, please read it.
Now these two bloggers are US-based - and this is significant because in the US there has been a huge emphasis on connecting between systems and exchange of document-based health information up until recent times. I view their postings as indicative of a growing trend toward the realisation that standardisation of clinical content is a necessary component for a successful health IT ecosystem in the (medium-longterm, sooner the better) future.
Note that "Detailed Clinical Models", is the current buzz phrase for any kind of model that might be standardised and shared but is also used very specifically for the HL7 DCMs currently in the midst of an interminable ballot process and the Australian national program's DCMs, which are actually openEHR archetypes being used as part of their initial specification process. "Detailed Clinical Models" is being used in many conversations rather blithely and with many not fully understanding the issues. On one hand it is positively raising awareness of our need to standardise content and on the other hand, it is confusing the issue as there are so many approaches. See my previous post about DCMs - clarifying the confusion.
It is worth flagging that there has been considerable (and I would also venture to say, rather premature) effort put in by a few to formalise principles for DCMs in the draft ISO13972 standard (Quality Requirements and Methodology for Detailed Clinical Models), currently out for ballot. My problem with this ISO work is that the DCM environment is relatively immature - there are many possible candidates with as many different approaches. It is also important to make clear that having multiple DCMs compliant with generic principles outlined in an ISO standard may mean that the quality of our published silos of "DCM made by formalism X" and "DCM made by formalism Y" models might be of higher quality, but it definitely will not solve our interoperability issues. For that you need a common reference model underpinning the models or, alternatively, a primary reference model with known and validated transformations between clinical model formalisms.
The more recent evolution of the CIMI group is really important in this current environment. It largely shares the principles that Kevin, openEHR and ISO13972 espouse - creation of standardised and shareable clinical content models, bound sensibly to terminology, as the basis for interoperability. These CIMI models will be computable and human readable; they will be based on a single Reference Model (yet to be finalised) and common data types (also yet to be finalised), and utilising the openEHR Archetype Definition Language (ADL) 1.5 as its initial formalism. Transformations of the resulting clinical models to other formalisms will be a priority to make sure that all systems can consume these models in the future. All will be managed in a governed repository and likely under the auspice of some kind of an executive group with expert teams providing practical oversight and management of models and model content.
Watch for news of the CIMI group. It has a influential initial core membership that embraces multiple national eHealth programs and standards bodies, plus all the key players with clinical modelling expertise - bringing all the heavy lifters in the clinical modelling environment into the same room and thrashing out a common approach to semantic interoperability. They met for 3 days recently prior to the HL7 meeting in San Antonio. The intent (and challenge) is to get all of this diverse group singing from the same hymn book! I believe they are about to launch a public website to allow for transparency which has not been easy in these earliest days. I will post it here as soon as it is available.
Maybe the planets are finally aligning...!
I have observed a significant change in the mind sets, conversations and expectations in this clinical modelling environment, over the past 5 years, and especially in the past 18 months. I am encouraged.
And my final 2c worth: in my view, the CIMI experience should inform the ISO DCM draft standard, rather than progressing the draft document based on largely academic assumptions about clinician engagement, repository requirements and model governance - there is so much we still need to learn before we lock it into a standard. I fear that we have put the cart before the horse.
Why the buzz about CIMI?
With the recent public statement from the Clinical Information Modelling Initiative (CIMI) my cynical heart feels a little flutter of excitement. Maybe, just maybe, we are on the brink of a significant disruption in eHealth. Personally I have found that the concept of standardising clinical content to be compelling and hence my choice to become involved in development of archetypes. During my openEHR journey over the past 5 or so years it has been very interesting to watch the changing attitudes internationally - from curiosity and 'odd one out' through to "well, maybe there's something in this after all".
And now we have the CIMI announcement...
So what has been achieved? What should we celebrate and why?
At worst, we have had a line drawn in the sand: a prominent group of thought leaders in the international health informatics domain have gathered and, through a somewhat feisty process, recognised that a collaborative approach to the development of a single logical clinical content representation (the CIMI core reference model) is a desirable basis for interoperability across formalisms. Despite most of the participants having significant investment and loyalty to their own current methodology and flavor of clinical models, they have cast aside the usual 'not invented here' shackle and identified a common approach to an initial modelling formalism from which other models will be derived or developed. Whether any common clinical content models are eventually built or not, naming of ADL 1.5 and the openEHR constraint model as the initial formalism is a significant recognition of the longstanding work of the openEHR Foundation team - the early specifications emerged nearly 20 years ago.
At its idealistic best, it potentially opens up a new chapter for health informatics, one that deviates from the relatively safe path of incremental innovation that we have followed for so many years - the reliance on messages/documents/hubs to enable us to exchange health information. There is an opportunity to take a divergent path, a potentially transformational innovation, where the focus is on the data itself, and the message/document/EHR becomes more simply just the receptacle or vehicle for the data. It could give us a very real opportunity to store lifelong health information; simplify data exchange (whether by messages or documents), aggregation, querying and analysis; and support knowledge-based activities such as decision support - all because we will (hopefully) have non-proprietary, common, agreed and fully defined models of clinical content and known transformations between each formalism.
Progress during the next few months will be telling. In January 2012, immediately before the next HL7 meeting in San Antonio, the group will gather again to discuss next steps.
There is a very real risk that despite best intentions all of this will fade away to nothing. The list of participating organisations, including high profile standards organisations and national eHealth programs, is a veritable Who's Who of international health IT royalty, so they will all come with their own (organisational and individual) work experience, existing modelling resources, hope, enthusiasm, cynicism, political agendas, bias and alliances. It could be enough to sink the work of this fledgling group.
But many are battle-weary, having been trudging down this eHealth path for a long time - some now gradually realising that the glacial incremental innovation is not delivering the long-term sustainable answers required for creating 21st Century EHRs as they had once hoped. So maybe this could be the trigger to make CIMI fly!
I think that CIMI is a very bright spark on the health IT horizon. Let's hope that with the right management and governance it can be agilely nurtured into a major positive force for change. And in the future, when its governance is mature and processes robust, we can integrate CIMI into the formal standards processes.
Best of luck, CIMI. We're watching!
CIMI & beyond...
The Clinical Information Modelling Initiative (#CIMI) is currently meeting in London. It comprises a significant group of healthcare IT stakeholders and was formed some months ago as an initiative by Dr Stan Huff. After a number of face to face meetings and email list exchanges, the intent is that at the end of this 3 day meeting there will be an agreed decision on a common clinical content modelling formalism/methodology for our Electronic Health Records. For background, from Sam Heard’s email to the openEHR email list on November 2, 2011:
The main topic I want to address is the international initiative to develop a standardised clinical modelling methodology. This has some IHTSDO secretarial support and is led by Dr Stan Huff of Intermountain Healthcare, a former HL7 Chairperson and co-founder of LOINC, who has been advocating a model-based approach for many years. The current approach at Intermountain has been influenced by openEHR and uses a two-level modelling approach. Stan has established a leadership group through trust and reputation, which includes a variety of agencies who have been working in the area and national eHealth programs or major initiatives who are interested in consuming the models. It has grown out of an HL7 Fresh Look initiative and is currently known as the Clinical Information Modelling Initiative (CIMI).
The group has committed to determining a single formalism for clinical modelling and ADL and openEHR are on the list of alternatives which is as follows:
- Archetype Object Model/ADL 1.5 openEHR
- CEN/ISO 13606 AOM ADL 1.4
- UML 2.x + OCL + healthcare extensions
- OWL 2.0 + healthcare profiles and extensions
- MIF 2 + tools HL7 RIM – static model designer
Proponents of the five different approaches have been presenting to members of the group, who have a variety of experience in these matters. Fourteen organisations will cast a vote on the formalism to use including openEHR, Singapore, UK NHS, Results 4 Care, HL7, Canada Infoway, 13606 Association, Tolven, CDISC, GE/Intermountain, US Departments, CDISC, SMArt and Mitre.
At the preliminary vote, held recently on November 20, the two most popular options were openEHR ADL 1.5 and UML.
Today CIMI will vote on a proposal for either ADL 1.5 or UML to be adopted as the initial common formalism for use, and determine a road map for coordinated development of semantically interoperable clinical models into the future. The potential impact of this is huge and exciting. It could be a disruptive change in health IT.
We hold our collective breath!
Don’t re-invent the (clinical content) wheel...
 It was with great interest that I read about the the recommendation for a universal exchange language in the recent release of the US report to the President: REALIZING THE FULL POTENTIAL OF HEALTH INFORMATION TECHNOLOGY TO IMPROVE HEALTHCARE FOR AMERICANS: THE PATH FORWARD.
I had asked the Direct project about the existence of a national plan for standardising clinical content only recently... It appeared that here was a plan after all.
It was with great interest that I read about the the recommendation for a universal exchange language in the recent release of the US report to the President: REALIZING THE FULL POTENTIAL OF HEALTH INFORMATION TECHNOLOGY TO IMPROVE HEALTHCARE FOR AMERICANS: THE PATH FORWARD.
I had asked the Direct project about the existence of a national plan for standardising clinical content only recently... It appeared that here was a plan after all.
So, to the report. The approach and benefits proposed started well...
The best way to achieve a national health IT ecosystem is to ensure that all electronic health systems can exchange data in a universal exchange language. The systems themselves could be designed in any manner desired — they could accommodate legacy systems that prevail or new recordkeeping systems and formats. The only requirement would be that the systems be able to send and receive data in the universal exchange language. (p41)
I have previously blogged about a universal health record underpinned by an application independent library of clinical content definitions, so the intent and benefits are well aligned with my preferred approach.
But then alarm bells started to ring....
Because of its multiple advantages, we advocate a universal exchange mechanism for health IT that is based on tagged data elements in an extensible markup language. If there were another equally good solution, it should also be considered; we have collectively been unable to think of one. (p43)
Issue #1: Isn't it more appropriate for step one to identify the need for standardised clinical content as a policy, rather than specify the format up front? Isn't that really the domain of health informatics experts as part of a subsequent work plan? I feel like we've skipped a couple of steps in the decision-making process. And are they really advocating the creation of this metadata-tagged XML from a zero starting point?
Issue #2: The last 9 words of that paragraph, "...we have collectively been unable to think of one." I'm glad that they are still open to equally good solutions being considered as indeed there are many ways that individuals, groups and organisations are exploring how to standardise clinical content definitions as the basis for a universal exchange mechanism.
In ISO TC 215, the International Standards Organisations Technical Committee for Health Informatics, there is a new work item which has been evolving for at least 2 years, although yet to attain committee draft status, known as ISO 13972 - Quality criteria for detailed clinical models. This work item is targeting a new international standard for determining quality criteria about the development of detailed clinical models - all clinical models, pick your flavour! In the world of international standards it has been recognised for years that with the plethora of different approaches to developing clinical models for EHRs, there is a need for some criteria to support quality aspect in their development. This work is being led by modellers from the Netherlands, with experts participating from the Australian, Danish, German, Swedish, US and Canadian standards organisations. Creating clinical content is definitely not a new field of endeavour by the time it enters the international standards arena.
So, I am extremely surprised that this expert PCAST group have not been able to 'collectively think' of an existing alternative.
In my last blog - Clinical Knowledge Governance in a Web 2.0 world – I pointed to a number of approaches to standardised clinical content to support health information exchange.
1. In the US – including, but by no means limited to:
- the HL7 standards organisation - where my UK colleague, Charlie McKay, informs me that there are more than 20 different approaches to clinical content development. Keith Boone (@motorcycle_guy) has posted his response to the PCAST report from a HL7 point of view - The Language of HealthIT;
- Stan Huff's group at Intermountain Health in Utah have had extensive experience in defining standardised clinical content across all of Intermountain's systems – they are leading experts in this domain; and
- I understand Don Mon and his team from AHIMA have also been working in this area.
2. In Europe, and Australia:
- The Australian National eHealth Transition Authority (NEHTA) has just launched a national clinician-led CKM approach to grassroots clinical content standardisation creating NEHTA's Detailed Clinical Models (DCMs).
- The Swedish national eHealth program has been actively working to integrate openEHR archetypes with its' VTIM architecture.
In addition, a few more points...
Firstly, the focus of the PCAST report is still only on data exchange, not on ensuring a sound foundation of a person-centric electronic health record. I'll say it again... get the data right and then the data will be able to be re-used, to multitask, be liquid, flowing to where it needs to be. It will become the solid foundation on which to build lifelong health records, simpler health information exchange, data integration & aggregation, research, reporting and knowledge-based activities. By focusing on exchange alone, then... you'll hopefully be able to exchange well and the rest will be considerably more uncertain.
Secondly, the proposed variant of XML is described as a 'straightforward' and 'superior' solution (p44), and the assumption that it will be scalable, protected by encryption, and that data element access services will be enough to support the health information exchange required. By contrast HL7, ISO/CEN 13606 and openEHR have taken decades to develop and refine underlying reference models to ensure that they have an unambiguous, consistent, secure way to represent personal health information – so you know who created the data, who is the subject of care, what the data means, what are the access rules applicable etc. In the openEHR environment, the specification authors developed Archetype Definition Language (ADL) for the purpose - and now part of the ISO 13606 standard - because the alternatives such as standard XML were not robust enough to represent health information. A 'straightforward' XML approach has a strong possibility of failure without a RM underpinning it.
And finally, there is the area of clinical knowledge governance itself. Health is dynamic, complex and diverse. The work required to represent healthcare as computable clinical content definitions or specifications is huge – don't underestimate the sheer volume of work that will be required. It is not realistic to expect a 'rapid mapping' of existing proprietary data structures into tagged data elements. Who will decide the clinical content in the models? If there are over 7000 clinical vendors in the US, which will be 'the source' or sources? Which are 'correct' or 'authoritative'? What methodology will be used to create the models? What level of granularity for each clinical element? How will they be aggregated together to represent clinical documents or events, and constrained to be useful for the clinical purpose? I have a million more questions...
Once the information models are defined, there will be a need for them to be validated before they can become the basis for a standardised or national clinical content library – suitable for consumers, clinicians, organisations, vendors, researchers and jurisdictions. A requirement will be recognised for life-cycle management and publication of these models, roadmaps for legacy data to migrate towards, and harmonise with, the new national health information 'source of truth', plus ongoing maintenance and governance.
Eric Browne stated in his recent blog, Recasting e-Health in the USA:
The work in Sweden, the UK, Singapore and even Australia, based on openEHR or ISO 13606 archetypes (i.e. implementable renditions of Detailed Clinical Models) is far more advanced and promising than that offered by the PCAST approach.
openEHR, which is my interest, has an approach to defining, agreeing and governing clinical content models for electronic health records, known as archetypes. It has taken more than 18 years to develop the openEHR technical specifications, and the last 10 years to achieve its' current approach and position in terms of clinical modelling. It is gaining traction, albeit with a modest volunteer community, especially now that it has a collaborative portal, known as the Clinical Knowledge Manager, to support sharing or models, reviews of clinical content, translation and terminology binding, and model governance.
Standardising health information definitions for health records or exchange is not a trivial task. Learn from what has already been achieved – all shapes, flavours and doctrines. Whatever you do, don't reinvent the wheel and create yet another universal language!
Clinical Knowledge Governance in a Web2.0 world
 Establishing and maintaining the quality of clinical knowledge is clearly the domain of the expert clinicians themselves. This is a broadly accepted principle for management and governance of the traditional clinical knowledge artefacts. However this assumption needs re-evaluation when we need to establish quality, safety and ‘fitness for purpose’ of computable clinical knowledge artefacts that populate Electronic Health Record (EHR) systems.
Clinical knowledge has traditionally been created and shared through formal publication and peer-review processes that have been adjudicated by committees of clinical experts. Those expert committees have been appointed through a credentialing process and have had jurisdiction and oversight over the entire publishable content – ‘the buck stops here’. Before the rise of the internet, face-to-face meetings have been where most of the committee work has been done, and the process has most often been slow and expensive but delivered good quality publications. The opportunity cost to each participating clinician has been high with recurring interruptions to their clinical activities. Revision of those publications at a later date repeats this process, taking considerable time, money and resources.
Establishing and maintaining the quality of clinical knowledge is clearly the domain of the expert clinicians themselves. This is a broadly accepted principle for management and governance of the traditional clinical knowledge artefacts. However this assumption needs re-evaluation when we need to establish quality, safety and ‘fitness for purpose’ of computable clinical knowledge artefacts that populate Electronic Health Record (EHR) systems.
Clinical knowledge has traditionally been created and shared through formal publication and peer-review processes that have been adjudicated by committees of clinical experts. Those expert committees have been appointed through a credentialing process and have had jurisdiction and oversight over the entire publishable content – ‘the buck stops here’. Before the rise of the internet, face-to-face meetings have been where most of the committee work has been done, and the process has most often been slow and expensive but delivered good quality publications. The opportunity cost to each participating clinician has been high with recurring interruptions to their clinical activities. Revision of those publications at a later date repeats this process, taking considerable time, money and resources.
Certainly in recent times, there have been more electronic tools to support these processes – email, teleconferences and videoconferences have improved the logistics of the process, but essentially the process remains unchanged.
Given the increasing traction of electronic health records, there is a parallel movement to develop and share computable clinical content definitions that can be created, published and implemented by: multiple clinical disciplines; generalists and specialists; primary, secondary and tertiary care organisations; population health planning; clinical researchers; and knowledge-enabled systems such as clinical decision support applications. They need to be language independent and translatable, in order to transport health information across national boundaries.
These kind of computable clinical models need the input from many experts, clinicians and others, to ensure that they are not only clinically appropriate but support safe data usage in our EHRs. These models are increasingly being created with ambitious goals – to create once and then re-use many times. In this case, the scope of the models needs to include requirements of the full breadth of clinical professions and specialties. Clinicians remain key to their development and publication, but they also require input from:
- Other domain experts – non-clinicians who will want or need to use these same models for non-clinical purposes such as secondary data use;
- Informaticians – who understand how these models will be the basis for recording health information, exchange between systems, reporting, data aggregation and how knowledge-based activities.
- Terminologists – to ensure that the models will integrate with appropriate terminology value sets;
- Technicians – who will advise on the technical impacts of these models in systems; and
- Translators – who will ensure that the clinical information is faithfully transformed from one language to another.
Examples of these computable clinical content models are many and varied. There are open source and proprietary models of many different flavours and philosophies – archetypes, templates, detailed clinical models etc. In recent years there are increasing attempts to broaden the input to the creation of these models and even to start to standardise them – regionally, nationally and even internationally. In this new paradigm, the traditional approaches to clinical content development, management and governance are no longer sufficient.
When the full breadth, depth, and dynamic nature of clinical knowledge is considered, it is not feasible to be able to appoint an overarching committee or board who would be capable of providing final ‘sign off’ about the clinical ‘correctness’ for any one model. Each clinical knowledge model will require input from varying groups of expert clinicians, terminologists, informaticians and technicians, depending on the clinical knowledge artefact under review. We need to find innovative approaches to online and asynchronous collaboration of a wide range of individuals from diverse backgrounds, expertise and geographical location to ensure these models are suitable for use in clinical systems.
Traditional standards bodies, such as ISO, CEN or HL7 have well defined and fixed processes in place for managing the lifecycle of technical standards through a formal balloting process with registered member bodies. These are definitely not suitable for managing and governing an evolving and dynamic clinical content specification library.
There has been some early work on establishing abstract archetype quality criteria by QREC and more recently, ISO TC 215 Working Group 1 has established a new work item 13972, which is establishing “Quality criteria for detailed clinical models”. However, neither of these are able to establish the quality of archetype instances for real world use.
I believe that HL7 is working to establish a Template Repository. As I understand it, it will operate as an indexing service to templates that will be stored on distributed servers. Others may be able to provide more details.
Other work is no doubt occurring, of which I am not aware. And of course, each clinical system has to establish the clinical content that it will use in its own proprietary information model. In the US alone, with thousands of clinical software vendors, this means that we have thousands of different computable versions of essentially identical clinical content, but none of it interchangeable without mappings or transformation – what a huge waste of resources! We need to change this blinkered way of thinking.
The openEHR Clinical Knowledge Manager (CKM) is the only online clinical knowledge resource, to my knowledge, which is supporting collaboration by clinicians, other domain experts, informaticians, technicians and translators to achieve consensus about quality and safety in clinical content models – in this instance, openEHR archetypes. I am directly involved in the development of this tool, and am active as an Editor facilitating the review process of the archetypes – I have described it in previous blog posts.
While CKM is one of the early Web2.0 approaches to collaborating about clinical content models, I am sure there will be more over time. I have spoken to a number of Knowledge Management experts, and to my surprise no-one has yet been able to point me to similar tools, resources establishing quality within a Web2.0 environment. Are we really such pioneers? Surely there are similar approaches in other knowledge domains?
No matter. There is no doubt that we are only in the early stages of a transformation in clinical knowledge governance and we have a lot to learn about how to establish quality criteria in a Web2.0 environment. I’ll post some thoughts in my next post...
Can clinicians agree?
 There have been a number of robust discussions in recent weeks around the claim that clinicians achieve consensus around computable definitions for clinical content. All discussions have been with MDs, some with substantial experience in various international standards organisations. All have been extremely sceptical...
There have been a number of robust discussions in recent weeks around the claim that clinicians achieve consensus around computable definitions for clinical content. All discussions have been with MDs, some with substantial experience in various international standards organisations. All have been extremely sceptical...
The first challenge: "What is a heart attack?” - and the response, “5 clinicians, 6 answers” – and they are probably very accurate!
The second: “What is an issue vs problem vs diagnosis?” - I'm told that this has been an unresolved issue in HL7 for 5 years+!
And the third, from an obstetrician: “The midwives want all this rubbish that we don’t” - perhaps an unfortunate way of expressing the absolutely correct need for different clinicians to have screens presented that are relevant to the patient and the immediate clinical task at hand. Different clinicians have different needs.
However these are all essentially HUMAN PROBLEMS! Issues about communications, synonyms, value sets, screen display/layout. IT will not solve these issues - as always, the clinicians need to work out the issues amongst themselves. So where CAN we achieve consensus?
Within the openEHR Clinical Knowledge Manager environment we are gaining some traction in achieving clinician agreement to the structure required to define clinical concepts as archetypes – the ‘first principles’ of clinical concepts, if you like. The approach is inclusive of everyone's needs and requirements, rather than requiring an arbitrary decision on the minimum data set or a priority data set - so we aim, as best we can, for a maximum data set.
For example, the framework to express a Diagnosis is largely not contentious:
- Diagnosis name
- Status
- Date of initial onset
- Age at initial onset
- Severity
- Clinical description
- Anatomical location
- Aetiology
- Occurrences
- Exacerbations
- Related problems
- Date of Resolution
- Age at resolution
- Diagnostic criteria
And for a Symptom:
- Symptom name
- Description
- Character
- Duration
- Variation
- Severity
- Current intensity
- Precipitating factors
- Modifying factors
- Course
- Aetiology
- Occurrences
- Previous episodes
- Associated symptoms
This notion of achieving clinician (and other domain expert) consensus and standardisation of clinical concepts is a major focus of the openEHR archetype work.
Bear in mind that while agreement can be achieved on the clinical content structure, this is only the first step in ensuring that clinicians are able to enter, retrieve and exchange meaningful clinical data.
So if we can achieve consensus around these archetypes, do clinicians then have to agree on a standard Discharge Summary or Antenatal Record or Report? The answer: only if is useful to do so. Clinical diversity can be allowed if the archetype pool is stable & governed/managed. By tightly governing the archetypes at international or national level, these are effectively the common 'lingua franca' that enables sharing of health information.
Template creation is the next openEHR layer - these aggregate the archetypes together to represent the requirements for a specific clinical activity and then constrain them down from their fully inclusive state to something that is 'fit for use' for a given clinician in a given clinical situation. Terminology subsets are also integrated in appropriate places into the archetypes (sometimes) and templates (usually) to round out the expressiveness needed by computable clinical models in clinical care - neither the structure nor the terminology can do this in isolation.
Once a critical mass of these archetypes are published we will be able to support a breadth clinical diversity across eHealth projects - the need for rigid, inflexible messages and documents to support any health information exchange has been largely overcome. Clinicians will only need to agree these types of messages or documents where there is a measurable benefit for doing so, and at all other times they can focus on ensuring that they express their archetype-based clinical records as flexibly as they need to for patient care. Sure, the human factors remain unresolved - but not even the most perfect EHR will solve these issues!
So can clinicians agree? Yes! It is happening in many areas related to data structure, creating a solid framework for our electronic health records. Archetypes are the tightly managed building blocks; templates enable safe and flexible expression of clinical and patient records - allowing the best of both worlds, both governance AND clinical diversity to flourish... and the clinicians are finally able to actively participate in shaping their EHRs.
Clinician-led eHealth records - a knowledge-enabled approach
My presentation given to W.H.O. in Geneva last week... [slideshare id=5492832&doc=20101008whoclinician-ledehealthrecords-101019133425-phpapp02]
Archetypes: the ‘glide path’ to knowledge-enabled interoperability
 In a world where connectivity is the universal aspiration, our health information is largely still caught up in silos and, in the main, is not accessible to those who need it – patients, clinicians, researchers, epidemiologists and planners. Shared electronic health records (EHRs) are increasingly needed to support the improvement of health outcomes by providing a timely, comprehensive and coordinated foundation for provision of healthcare. For decades people have been attempting to share health information, but the incremental approach has not been wholly successful – progress has been made, but despite enormous investment and resources, the solution has been found to be more difficult than most anticipated; many well-funded attempts have been stunningly unsuccessful. Healthcare provision appears not to fit the model that has been so successful in other domains such as banking or financial services. Why has sharing health information been so difficult? After all, on the surface, data are, simply, data.
In a world where connectivity is the universal aspiration, our health information is largely still caught up in silos and, in the main, is not accessible to those who need it – patients, clinicians, researchers, epidemiologists and planners. Shared electronic health records (EHRs) are increasingly needed to support the improvement of health outcomes by providing a timely, comprehensive and coordinated foundation for provision of healthcare. For decades people have been attempting to share health information, but the incremental approach has not been wholly successful – progress has been made, but despite enormous investment and resources, the solution has been found to be more difficult than most anticipated; many well-funded attempts have been stunningly unsuccessful. Healthcare provision appears not to fit the model that has been so successful in other domains such as banking or financial services. Why has sharing health information been so difficult? After all, on the surface, data are, simply, data.
Why is the health information domain different?
Health information is the most multifaceted and largest knowledge domains to try to represent in a computer. The SNOMED CT terminology alone has over 450,000 terms expressing health-related concepts, and our collective knowledge about health is far broader, deeper and richer than that required to represent financial systems. The added bonus in health is that our information domain is dynamic - growing and changing as our understanding increases.
Recording, communicating and making sense of health information is something that clinicians do remarkably well in a localised, non-digital world. However the human cognitive processes and assumptions that underpin the traditional health records do not easily translate into the computerised environment. Consider the need for narrative versus structured data; the complexity of clinical statements; use of the same data in a variety of clinical contexts; the need for clinicians to make ‘normal’ or ‘nil significant’ statements, and also the oft underrated positive statements of absence; and the need for graphs, images or multimedia in a good health record. Grassroots clinicians have different personal preferences for creating their clinical records and to support their requirements for direct provision of clinical care and communication to colleagues. In parallel, jurisdictions have different expectations of the grassroots clinical data collection that will support reporting, data aggregation and secondary use of data.
Throw into this mix the complex and convoluted processes required to support healthcare provision; mobile patient populations; and the need for lifelong health records, and it starts to become easier to understand why eHealth has been more of a challenge that many first thought.
Information-driven EHRs
Traditional approaches to the development of EHRs have been software application-driven, hard-coding clinical knowledge into the proprietary data model for each software system and resulting in silos of health information locked away in proprietary databases. This is valuable data, and even more valuable if we can get access to it, exchange it and utilise it. Key stakeholders – patients, clinicians, researchers, planners and jurisdictions – are currently disempowered and are not easily able influence or express their data requirements. We have mistaken the software application for the electronic health record - a classic example of the ‘tail wagging the dog’.
If we focus on the electronic health record being the data, we turn the traditional paradigm upside down. Our EHRs become information-driven by putting the stakeholders at the centre to direct the information content and quality aspects of our EHR systems. It is only then that our systems will be able to reflect the real requirements of stakeholders, ensuring that health information collected data is ‘fit for use’ and will support personal health records, clinician health records and, with appropriate authorisation and permissions, the broadest range of secondary use.
Sharing health information requires common and coherent health information definitions or models – ensuring that health information can be expressed in a way that is meaningful to stakeholders AND that computers can process it. According to Walker et al , Level 4 interoperability, or ‘machine interpretable data’, comprises both structured messages and standardised content/coded data. In practice, it means that data can be transmitted and viewed by clinical systems without need for further interpretation or translation. This semantic, or knowledge-level, interoperability is absolutely required for truly shareable health records, data aggregation, knowledge-based activities such as clinical decision support, and to support comparative analysis of health data. Further, it is only when this health information model is agreed at a local, regional, national or international level, that true semantic interoperability can occur at each of these levels. The broader the level of clinical content model agreement, the broader the potential for semantic health information exchange.
The openEHR paradigm
openEHR is a purpose-built, open source, information-driven electronic health record architecture focused on ensuring that the grassroots health information is recorded clearly, coherently and unambiguously in EHRs, and supporting re-use in other contexts where appropriate. It adopts an orthogonal approach to EHRs - a dual-level modelling methodology with clear separation of the technical from the clinical domains, where software engineers focus on their application development and the clinical domain experts focus on the health information definitions. openEHR focuses on the data - using computable knowledge artefacts known as archetypes and templates to formally express health information.
openEHR archetypes are computable definitions created by the clinical domain experts for each single discrete clinical concept – a maximal (rather than minimum) data-set designed for all use-cases and all stakeholders. For example, one archetype can describe all data, methods and situations required to capture a blood sugar measurement from a glucometer at home, during a clinical consultation, or when having a glucose tolerance test or challenge at the laboratory. Other archetypes enable us to record the details about a diagnosis or to order a medication. Each archetype is built to a ‘design once, re-use over and over again’ principle and, most important, the archetype outputs are structured and fully computable representations of the health information. They can be linked to clinical terminologies such as SNOMED-CT, allowing clinicians to document the health information unambiguously to support direct patient care. The maximal data-set notion underpinning archetypes ensures that data conforming to an archetype can be re-used in all related use-cases – from direct provision of clinical care through to a range of secondary uses.
Templates are used in openEHR to aggregate all the archetypes that are required for a particular clinical scenario – for example a consultation or a report. These can also be shared, preventing more ‘wheel re-invention’. Individual content elements of each maximal archetype can be ‘disabled’ in the template so that the only data elements presented to the clinician are those that conform to national or local requirements and are relevant and appropriate for that use-case scenario. For example, a typical Discharge Summary may commonly comprise 10 common archetypes; templates allow the orthopaedic surgeon to express a slightly different ‘flavour’ of the Discharge Summary based on which elements of each archetype being either active or disabled, compared to that required by a Obstetrician who needs to share information about both mother and newborn. One ‘size’, or document, does not fit all. The archetypes, as building blocks, are the key to semantic interoperability; while templates allow flexible expression of the archetypes to fulfill use-case requirements.
How achievable is this? Only ten archetypes are needed to share core clinical information that could save a life in an emergency or provide the majority of content for a discharge summary or a referral. If each archetype takes an average of six review rounds to reach clinician consensus and each review round is open for 2 weeks, it is possible to obtain consensus within an average of three months per archetype – some complex or abstract ones may be longer; other simpler, more concrete archetypes will be shorter. Many archetypes are already well developed in the international arena and within national programs. As archetype reviews can be run in parallel, a willing community of clinicians could achieve consensus for core clinical EHR content within three to six months.
It is estimated that as few as fifty archetypes will comprise the core clinical content for a primary care EHR, and maybe only up to two thousand archetypes for a hospital EHR system including many clinical specialties. The initial core clinical content will be common to all clinical disciplines and can be re-used by other specialist colleges and interested groups. More specialised archetypes will gradually and progressively be added to enhance the core archetype pool over time.
The openEHR Clinical Knowledge Manager (CKM) is an online clinical knowledge management tool – www.openEHR.org/knowledge - which provides a repository for archetypes and other clinical knowledge artefacts, such as terminology subsets and document templates. Based on a data asset management platform it provides a clinical knowledge ecosystem supporting the publication lifecycle and governance of the archetypes. Within CKM, a community of grassroots clinicians and health informaticians collaborate in online reviews of each archetype until consensus is reached and the agreed archetype content is published. Clinicians and other domain experts need no technical knowledge to engage with archetypes - the technical aspects of archetypes are kept hidden ‘under the bonnet’ – but they use their expertise to ensure that the content definitions within each archetype is correct and appropriate. Each content review is conducted online at a time of convenience to the clinician and usually only takes five to ten minutes for each participant. Thus the clinical domain experts themselves drive the archetype content definitions, and CKM has become a peer-reviewed knowledge resource for all parties seeking shared, standardised and computable health information models.
At the time of writing CKM has acquired, largely by word of mouth, 565 registered users from 62 countries, including 181 people who have volunteered to review archetypes, and 73 who have volunteered to translate archetypes. The repository contains 273 archetypes, of which 15 have content that are in team review and 9 published. Two example templates have been uploaded, and we await final publication of the openEHR template specification before we expect to see template activity increase. Terminology subset functionality has been added only recently and our first terminology subsets uploaded. So, while CKM is still relatively new, its Web2.0 approach to artifact collaboration and publishing, combined with formal knowledge artifact governance positions CKM as a pioneering ‘one stop shop’ for clinical knowledge resources online.
Current CKM functionality includes:
- Display of artefacts including structured views, technical representations and mind maps to make it easy for clinicians and others to review;
- Uploading of new knowledge artefacts – archetypes, templates and terminology subsets – for review and publication;
- Archetype metadata supporting classification, ontological relationships and repository-wide searches;
- Digital asset management including provenance and artefact audit trail;
- Integration with openEHR tools supporting quality assessment & technical validation checks;
- Review and publication process for clinical content – draft, team review, published and reassess states
- Terminology binding and terminology subset reviews;
- Online archetype translation with review;
- Community engagement via threaded discussions, repository downloads, attached resources, watch lists, email notifications, user dashboards and release sets;,
- Editorial support via To Do lists, user and team administration, review management, artefact modification, classification management, broadcast emails etc;
- Subscriber auto-notification including Twitter and email
- Reports – Archetypes, Templates and Registered users
A governed repository of shared and agreed archetypes will provide a ‘glide path’ towards full semantic interoperability of health information; a clear forward path for standardisation of data definitions. These will bootstrap new application or program development, provide a ‘road map’ to support gradual transition of existing systems to common data representation and provide the means to integrate valuable silos of legacy data.
Benefits of a collaborative, data-driven approach
A collaborative and domain expert-led approach to our health information provides many benefits which include the following.
Benefits for stakeholders
- Active involvement of domain experts to ensure the safety and quality of health information.
- Development of a coherent set of health information definitions:
- Improved data quality – shared core clinical content plus specialised domain-specific content will be agreed and ratified by the domain expert community; health information created will need to conform to the agreed archetype specifications.
- Improved data ‘liquidity’ – specifications to support exchange, flow and re-use of health information - from direct patient care through to secondary use of data. Improved data longevity – shared non-proprietary health information definitions minimise need for data transformations or system migration and the inherent risk of data loss; will support the cumulative, lifelong health records and longitudinal data repositories;
- Improved data availability – easier integration of health information from disparate sources when based on common archetype definitions;
- Re-use, integrate and aggregate data for supporting quality processes such as clinical audit, reporting and research; and
- Break down the existing ‘silos’ of health information based on proprietary and varied definitions.
- Online collaboration maximises the potential for a breadth of grassroots stakeholder engagement in ensuring correctness of the health information definitions.
- Active participation by clinical domain experts to shape and influence their EHRs, ensuring that EHR content is ‘fit for clinical purpose’.
- Online participation in clinical content review will be of short duration and at times of convenience to the clinician, avoiding the significant time and opportunity cost of attendance at face-to-face meetings.
Benefits for patients
- Data created and stored in a shared, standardised and non-proprietary representation supports the potential for application-independent data records that can persist for the life of the patient.
- Improved data ‘liquidity’ – so that data can flow between healthcare providers and systems to where the patient needs it.
Benefits for national programs and other jurisdictions
- Development of a coherent national set of clinical content specifications to support the shared EHR programs, health information exchange and secondary use.
- Enables national governance of foundation clinical content while at the same time facilitates flexible expression of local domain requirements
- Efficient use of sparse clinical, informatics and stakeholder resources:
- Design & create an archetype once; re-use many times;
- Leveraging existing clinical specification work done internationally to improve local national pool of archetypes;
- Online collaboration maximise the potential for stakeholder engagement at the same time as minimising the requirement for expensive face-to-face meetings; and
- Review and publication of agreed clinical specification definitions within weeks to months;
- Review and standardisation of clinical documents containing agreed archetypes will be relatively short.
- • Clinical knowledge management ecosystem:
- Single national repository of clinical knowledge artefacts, including archetypes and terminology subsets.
- Focussed and coordinated knowledge management environment where all stakeholders can observe, participate and benefit; the opposite of the current fragmented, isolated and proprietary approach to defining health information content.
- Digital knowledge asset management:
- Manages authoring, reviewing, publication and update lifecycle of all knowledge assets;
- Provenance and asset audit trails;
- Ensures asset compliance to quality criteria;
- Ensures technical validation of assets; and
- Development of coherent release sets for implementers;
- Governance of knowledge assets.
- Distribution of knowledge assets via coherent release sets.
- Removes the need for per message or per document negotiation between application developers, organisations and jurisdictions each time information needs to be integrated or exchanged by use of the standardised content within more generic message wrappers or document structures.
- Transparency of editorial and publishing processes; accountability to the domain expert community itself.
- Precludes the need for ratification of clinical or reporting documents through a traditional standards process when they consist of subsets of the nationally agreed archetypes.
Benefits for application developers
- Download coherent sets of clinical content definitions from a published and agreed national repository – not re-inventing the wheel by defining each piece of health information over and over.
- Software development remains focused within the expert technical domain – user interface; workflow processes; security, data capture, storage, retrieval and querying; etc.
- Removes the need for per message or per document negotiation between vendors, organisations and jurisdictions each time information needs to be integrated or exchanged by use of the standardised content within more generic message wrappers or document structures.
Benefits for secondary users of data
- Existing data can be mapped to archetypes once only, and transformed into a validated and consistent format; new data can be captured and aggregated according to the same national archetype definitions.
- Data stored in a common representation can be more easily aggregated and integrated.
- Access to valuable legacy data that would otherwise be unavailable.
Agreed and shared representations of the health information, embracing existing stakeholder requirements and developed rapidly by an active online community, will kick-start and accelerate many currently fragmented eHealth activities. Sharing archetypes as the definition of our health information will not only provide a common basis for recording and exchanging health information but also simplify data aggregation of data, support knowledge-based activities and comparative data analysis. Perhaps even more compelling, we are making certain that our domain experts warrant that the data within our EHRs, and flowing between stakeholders, is safe and 'fit for purpose'.
What if... grassroots professional clinical colleges were driving EHRs?
Is the tail wagging the dog? Should vendors decide what clinicians need? OR should the professional clinical colleges be driving the EHRs that members need to provide quality healthcare to their patients? From a quality point of view, one could strongly suggest that colleges should be the experts driving this new paradigm of clinical care. As a group, professional clinical colleges as a group usually state that one of their major roles is in establishing and driving the benchmarks for acceptable clinical practice standards in their field of expertise. Most do this very well in the traditional areas of clinical practice.
Scouring the web it is possible to see that some are engaging actively in eHealth. For example, from the website of The Council of the Royal College of Physicians in UK, a vision statement published in January 2010 incorporates this final paragraph:
"Effective implementation of standardised, structured, patient focused records requires strongly led culture change, embraced by all medical and clinical staff. They are essential prerequisites for safe, high quality care and for the safe, efficient and effective migration from paper to electronic patient records. Such records will also enable innovative development of services that cross traditional boundaries and, by giving patients access to their record, empower them to take more responsibility for their own care."
If colleges have had a traditional role in ensuring safe, high quality care, then the advent of electronic health information should logically trigger a natural extension of their pre-existing work and roles into the eHealth arena. I would go further and suggest that they should also be actively driving an eHealth agenda that reflects their college remit regarding provision of quality care and standards; ensuring that the data created, stored, shared and queried is good and fit for use in clinical care.
This agenda may, or may not, be aligned with the activities of national eHealth programs. As we look at the news in recent weeks it is becoming apparent that many are struggling with implementation of top-down agendas. We have seen major changes in the national approach to eHealth happening in Netherlands, Germany, and England's NHS. Perhaps it is time for a grassroots, bottom-up approach from the end-users, the healthcare providers, and their patients, the healthcare consumers.
How can colleges drive eHealth?
- Promote the establishment of a universal health record - a long-term, data-driven health record platform based upon open source specifications, a standardised health data structure, and application-independence. If we place the patient's universal health record at the focus, applications can effectively 'plug & play' with an open, standardised patient data repository, instead of struggling/battling to move/transform/migrate/map/message bits of health information between proprietary application silos.
- Develop clinical quality-related eHealth standards:
- Knowledge artefacts specifications The clinical knowledge expertise of colleges can be used not only to develop principles of best practice and clinical guidelines etc but also the structured clinical content specifications that underpin the universal health record and ensure that the health record is able to capture all the data that their clinicians need to provide quality clinical care, to share with other healthcare professionals and for research. These specifications need to be defined both at the clinical concept level, such as the specification for blood pressure, but also expressing how many clinical concepts can be aggregated together and constrained to specify the requirements for a computable discharge summary or an anaesthetic record. Ideally this would be work done in collaboration with other clinicians, colleges and informaticians to make sure that the agreed concept specifications are applicable for all colleges and clinical domains, so that there is one common concept in use for all. Colleges as domain experts are also best placed to determine the terminology subsets that are appropriate for use by their clinicians.
- EHR best practice: Develop principles around the best practice use of electronic health records, including:
- Data quality principles and activities. As health records become electronic, the previous remit of Colleges to make sure that clinical practices are accredited and that paper records are kept to a documented standard can be transformed to the eHealth paradigm. There are similar principles that can be developed and applied when clinicians are capturing and using data. Programs such as Primus Plus in the UK have worked directly with primary care clinicians to educate and support about best practice data management.
- EHR safety - User interfaces,clinical processes, decision support etc
Colleges may bundle these new standards-based eHealth activities within existing service provision to, or activities involving, their members.
In addition, commercially savvy colleges could develop ways to develop and sell practical solutions or packages around these quality principles direct to members – products or programs to make it easy for members to put the evidence-based and quality principles into practice.
Herein lies a challenge for contemporary professional clinical colleges – to both embrace eHealth and to actively drive it in a way that promotes and preserves safety, quality and best practice in healthcare, such that eHealth becomes a positive tool for healthcare provision and, possibly even, reform.
And how to do this in practice? Well, it is no secret that I think openEHR can offer a solution to a lot of these issues - see my previous posts 'What on earth is openEHR' and 'Connect with openEHR'.
Time will tell if I'm on the right track. For me as a clinician, the more time I spend with it, the more compelling this openEHR story becomes...
What on earth is openEHR?
 My experience in eHealth started as a suburban General Practitioner using an EHR application for prescribing and clinical notes, and then I moved sideways, becoming involved in building proprietary clinical software and a Personal Health Record. From 2000 I worked for 4 years in a single company that owned that PHR plus a primary care clinical application and a hospital application - the intent being that data created in one could be transferred between the systems, but we found it wasn't that easy - for various reasons they all had different database structures, even within the same vendor! So if we have to do the same thing between disparate vendors in an environment that is more competitive than collaborative, the picture becomes infinitely more complicated.
In more recent years, I have had my world view shifted from the traditional application-drive EHR to a data driven health record (note the deliberate lack of capitalization) - see my previous blog posts. Once we focus on getting the data right, capturing it or displaying it in the applications are just one of the many things we can do with the data.
My experience in eHealth started as a suburban General Practitioner using an EHR application for prescribing and clinical notes, and then I moved sideways, becoming involved in building proprietary clinical software and a Personal Health Record. From 2000 I worked for 4 years in a single company that owned that PHR plus a primary care clinical application and a hospital application - the intent being that data created in one could be transferred between the systems, but we found it wasn't that easy - for various reasons they all had different database structures, even within the same vendor! So if we have to do the same thing between disparate vendors in an environment that is more competitive than collaborative, the picture becomes infinitely more complicated.
In more recent years, I have had my world view shifted from the traditional application-drive EHR to a data driven health record (note the deliberate lack of capitalization) - see my previous blog posts. Once we focus on getting the data right, capturing it or displaying it in the applications are just one of the many things we can do with the data.
Why? I first heard about openEHR nearly 10 years ago. I didn't understand openEHR at all initially, but there was something in the commonsense of getting the foundation data defined and standardized that resonated with me. Over time I have become convinced that openEHR provides an orthogonal approach to eHealth that has a very reasonable chance of success, and more importantly, of making a difference. I no longer believe that the traditional application-driven approach to electronic health information management is effective, economic or sustainable.
 What is openEHR?
What is openEHR?
Think of openEHR as the open source health equivalent of the iPod/iPhone platform – a technical framework which will allow any compatible application, organization or provider to share 'plug and play' access to standardized data. This is openEHR’s innovation – the focus on ensuring that the underlying health data is correct, robust and trustworthy!
Rich health data definitions known as archetypes, are defined and agreed by the clinicians themselves to ensure that each piece of health information is unambiguously understood, ‘fit for purpose’ and can be dynamically used & reused to support wise and safe health choices, now and into the future. These same archetypes are also computable, so that when these common data definitions are shared, they act as a 'lingua franca', making it much simpler to capture, store, aggregate, query and exchange health information - effectively making the data 'sing and dance' and to flow according to privacy and access rules.
Developed over more than 15 years through international research, community input and implementations, openEHR is purpose-designed as a non-proprietary universal health record: application independent, yet supporting accurate and safe health information exchange between software programs, consumers, health care providers, organisations and researchers; and across the diverse requirements of private/public providers and regional, national and international jurisdictions.
Why openEHR?
- It is open source - break down the proprietary silos of data created by application vendors.
- It is the basis for the recently published ISO13606 standard for EHR extract. openEHR evolved and grew away from 13606 approx. 5 years ago as 13606 entered the CEN and ISO standards approval process; openEHR has subsequently progressed and developed as a direct result of implementation experience. At present openEHR is the commonest implementation pathway for nations mandated to adopt the 13606 standard.
- It is driven by an international open source community.
- It has been developed using a robust engineering process.
- The clinical content is driven by the domain experts - usually, but not limited to the clinicians themselves - through the Clinical Knowledge Manager.
- Archetypes are designed as maximal data sets for the universal use-case so the same data definitions can be used in any software application - whether a PHR, EHR, research project, clinical decision support system or running population queries.
- It is purpose-designed as a shared health record.
- The structured archetype definitions complement other standards-related work - for example the recent announcement of a collaborative work program between the openEHR Foundation and IHTSDO to explore how the SNOMED-CT and openEHR archetypes can be combined to provide a strong semantic solution for health information.
Who is using openEHR?
International momentum is building - current users noted on the openEHR website include commercial, government, academic, and non-profit organisations.
___________________
Current eHealth developments are progressing at a glacially slow rate because we are trying to develop interoperability by traditional and incremental methods. I'm increasingly sceptical that this is effective, economic or sustainable. And in fact, though direct experience I have become convinced that openEHR's orthogonal approach can make a significant difference. I now work with openEHR every day, including:
- alongside an international group of like-minded clinicians collaborating in their spare time in a Web2.0 application to develop, publish and govern the archetypes;
- with national eHealth programs who are seeking to build a library of clinical content definitions as a 'single source of truth' to mandate for use by vendors;
- with vendors who no longer have to reinvent the wheel but can take the published archetypes and re-use them within their systems; and
- with researchers trying to aggregate and integrate disparate data from multiple sources into one cohesive repository on which they can query their valuable data.
I'm not trying to sell you a software application like everyone else claiming an 'eHealth solution'; I'm trying to persuade you to take a look at an alternative approach to eHealth, to share a little of my vision and my passion!
Consider becoming part of an international open source community. Contribute and collaborate alongside other clinicians, informaticians and techies to build something bigger than all of us. Share a vision of how eHealth could work better and more effectively if we get the foundations for a universal health record consistent and solid.
It. is. all. about. standardizing. the. data.
Standardized, data-driven eHealth
 What is an EHR? Most currently working within eHealth circles will have a ready answer, and the classic response will be something like "an EHR is a software application that is used by clinicians for provision of patient care".
But think about it for a second...
What is an EHR? Most currently working within eHealth circles will have a ready answer, and the classic response will be something like "an EHR is a software application that is used by clinicians for provision of patient care".
But think about it for a second...
What do we want from an electronic health record? It's not actually the application or the user interface or the workflow, but a record of health information in an electronic format.
And so what of the health information itself? What is it? What do we want? What should we want?
Let's work with these definitions: Data are the computable facts; while health information is the data after it is processed, organized, structured or presented in a given context so as to make it useful.
We definitely want health information that...
- Is available at the right time and the right place to the right person.
- Is accurate, detailed and of high quality.
I don't think there is much disagreement on these two points. How about these next suggestions?
We should also want health information that...
- Is a lifetime health record - cradle to grave. Health information that can be accumulated over time, aggregated from many sources to inform better care for ourselves, our patients and loved ones.
- Can be created in one place or by one provider and shared with others so that the meaning of the data is preserved and accurate.
- Can be dynamically and actively used - consisting of structured, atomized data that we can re-use and combine in different ways to improve our health and wellness. Documents or PDF's of our health information are useful as part of a passive record, but they are effectively a dead end - we can't do anything useful with big blobs of text except read them.
- Is not locked in to a proprietary vendor database; data should be in an open, non-proprietary format.
- Is a continuum of our health information across all aspects of healthcare and related activities, not fragmented silos of data artificially segmented for personal, clinical or secondary use purposes.
- Can be accessed via an EHR, EMR or PHR, SEHR or ICEHR - pick an acronym, any acronym!
- Can be accessed independent of any specific organizations, providers and geographical locations.
- Has had minimal transformation or manipulation. Keep in mind the Chinese Whispers effect! There is significant risk that important data can be lost or lose integrity with each data migration to a new software application or transformation between disparate systems.
It is a commercial reality that we continue to develop EHR applications in the traditional manner, building the greatest and 'best of breed' clinical software applications, with each EHR vendor doing it in their own proprietary way, as 'rugged individuals'! The resulting software usually has a rich functionality and a great user interface. It is likely that it does a fine job locally in the clinic, hospital or network on which it is installed. But what about regionally, nationally, internationally? Is it still working well if we take the big picture view? Sure, our EHR applications are full of data, but the data is isolated, fragmented, and limited in its use.
There is no doubt that all over the world, the health IT community are finding it unexpectedly hard to exchange health information between different applications - the effort and resources that are being poured into policy, research and pilots for health information exchange is enormous. Progress is glacially slow. If we want to exchange PDFs and documents, then we are doing well, but if we want our software to be able to do more than display text, to do clever things with our health information, then our data requirements are much more complex. If we want that data to be able to be shared, used in multiple applications then the current approach requiring data transformations and messaging paradigms won't be sustainable
Interestingly, the further we go down these paths many are realizing the need to change our emphasis away from the EHR application and towards the data. Back in November 2009, Clay Shirky wrote:
"This ability to separate data from transport and applications from data is the essential pre-condition for innovation — a group that has a valuable new idea for presentation of data for clinical use should not also be forced to think about the data encoding or the way the data are transported. Groups working on new data encodings should not be tied to a pre-existing suite of potential applications, nor should they have to change anything in the transport layer to send the new data out, and so on."
The bolded text, above, reflects my emphasis on an important statement. Confirming this approach, as recently as this week, Kibbe & Klepper also called for separation of the data from the applications and from the transport layer.
Change the focus to standardized, data-driven eHealth
So, innovation requires a new approach to data; a changing emphasis from application- or message-driven to data-driven eHealth. If we also insist on an open and standardized approach to health data specifications then we will be able to realize many additional advantages:
- A strong foundation of shareable and re-usable computable clinical content definitions on which to build coordinated and cohesive applications, messages, clinical decision support programs, and research activities. If we use common, standardized data definitions then the tasks of eHealth become orders of magnitude easier. Content definitions are created and agreed as the content is already specified and the processes become more generic
- An unambiguous and detailed understanding of what each piece of data means so we can do 'stuff' with it - a tight semantic 'handle'.
- A powerful enabler for managing the complex requirements involved in health data capture, integration, aggregation, inferencing and sharing.
- A continuum of our health information, independent of vendor, provider or organization - the real potential for lifelong health records, for the first time.
There is no doubt that this approach is orthogonal to the status quo and it will be challenging to many for logistical, financial and political reasons, but can we really afford to ignore this?
According to the European Commission's seminal 2009 report entitled "Semantic Interoperability for Better Health and Safer Healthcare: Research and Deployment Roadmap For Europe" (PDF, p16), standardizing the capture, representation and communication of clinical data requires three components to represent meaning: a generic reference model for representing clinical data, agreed clinical data structure definitions and clinical terminology systems. Potential standardized data definitions proposed are openEHR archetypes, ISO/EN 13606 Part 2 (which are simpler archetype structures), HL7 templates, generic templates and data sets. Standardization of data is not 'pie in the sky' but an approach that has had significant research and implementation experience, particularly in Europe.
So, to consumers, clinicians, organizations, researchers and governments, the call should not only be "Gimme my damn data!" but give me standardized data that is application- and message-independent. Then we can actually start to use and re-use our data, not only as a detailed record of current and past health conditions, events and activities, but dynamically and pro-actively to inform and promote our future health and well-being.
PHRs: still a powerful grassroots approach to eHealth
 In 1999 four clinicians and an ex-CIO gathered around my kitchen table and week by week we hatched a plan for a Personal Health Record (PHR) which we hoped could revolutionise healthcare. At that time a PHR was not at all mainstream. Very few clinicians we spoke to thought it was a good idea at all. Many held a very paternalistic view that patients were not capable to handle their health information, and that a PHR would not ever be trusted as a useful resource (- for some reason a verbal a history was acceptable but a patient entered record of some sort was not)! Patients were also pretty passive and had to be convinced that they could be supported to manage their health information in a similar way that they managed their finances!
In January 2000, those last heady days of the dot-com era we started development in earnest, then rode the rollercoaster and navigated the fallout of the dot-com crash. HotHealth
In 1999 four clinicians and an ex-CIO gathered around my kitchen table and week by week we hatched a plan for a Personal Health Record (PHR) which we hoped could revolutionise healthcare. At that time a PHR was not at all mainstream. Very few clinicians we spoke to thought it was a good idea at all. Many held a very paternalistic view that patients were not capable to handle their health information, and that a PHR would not ever be trusted as a useful resource (- for some reason a verbal a history was acceptable but a patient entered record of some sort was not)! Patients were also pretty passive and had to be convinced that they could be supported to manage their health information in a similar way that they managed their finances!
In January 2000, those last heady days of the dot-com era we started development in earnest, then rode the rollercoaster and navigated the fallout of the dot-com crash. HotHealth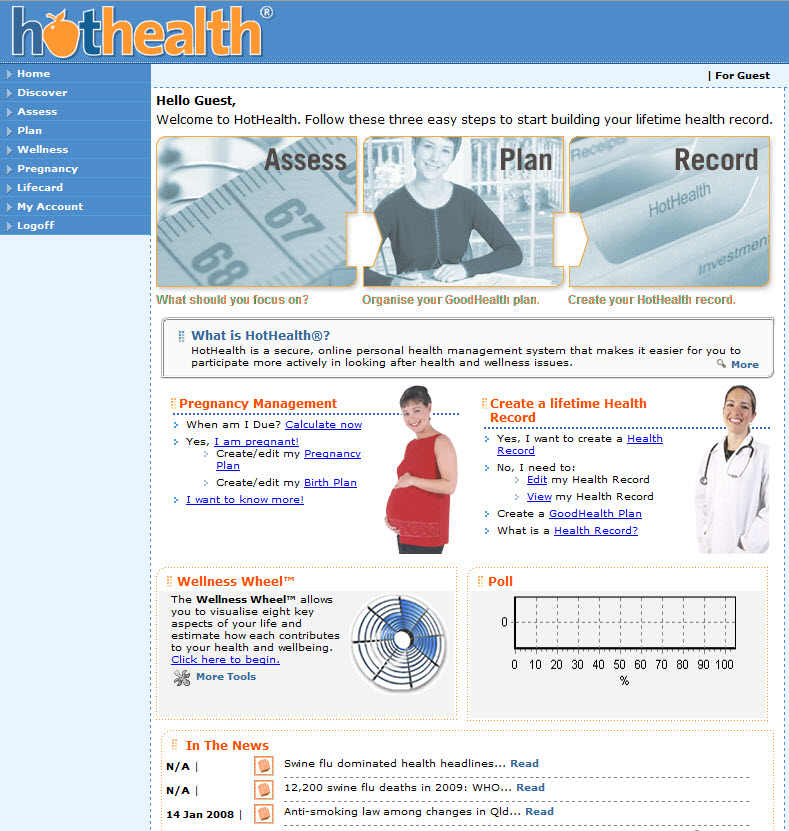 was launched in November 2000. My clinical colleagues were still not very enthusiastic about the concept. The business model was not easy. Large companies, health insurers, hospitals, pharmaceutical companies - everyone could see how a PHR would be 'useful', but none would commit. The funding dried up and it/we were acquired by listed company in 2002.
was launched in November 2000. My clinical colleagues were still not very enthusiastic about the concept. The business model was not easy. Large companies, health insurers, hospitals, pharmaceutical companies - everyone could see how a PHR would be 'useful', but none would commit. The funding dried up and it/we were acquired by listed company in 2002.
In the US, there were a fair number of PHRs in 2000, maybe even as many as 50. DrKoop.com (now apparently HealthCentral) was the clear leader at the time; WebMD is probably the only significant one remaining. Our annual market research for competitors showed a huge turnover - PHRs were appearing and disappearing at a great rate!
But the evidence was starting to come in and support the concept - Kate Lorig's work at Stanford on supported self-management in arthritis was working well on paper and face-to-face. We applied some of these principles to HotHealth plus health summaries, prevention plans, wellness promotion, quality health information etc - the kinds of features we are used to seeing in PHRs now.
Well, HotHealth still exists and has had some modest success, mostly in the spin off of a PHR for older children and teenagers with insulin dependent diabetes which ran between 2003 and 2005 - Betterdiabetes - but it never blossomed as I had hoped. Was it the timing? Was it the business model? Was Australia too small a market. Yes to all of these, and much more - it was harder than we anticipated.
- but it never blossomed as I had hoped. Was it the timing? Was it the business model? Was Australia too small a market. Yes to all of these, and much more - it was harder than we anticipated.
In 2005 I was asked to write a commentary in Australian Health Review - "The patient’s memory stick may complement electronic health records"(PDF of full text). Re-reading it now, I can't help but be disappointed that not a lot has really changed in terms of integration and data exchange between clinicians and patients. We have made some small progress, but I thought the revolution would have happened by now.
Amusingly I think that my final paragraph in that commentary, written back in September 2005, still stands true.
"A grassroots push by consumers wishing to hold, own and manage their own health information has the potential to make relatively quick, inexpensive and significant changes to the way healthcare is delivered, to support and encourage consumer input to their own health information record, and kick-start electronic health record development in Australia. And eventually when our individual PHRs evolve to amalgamate and integrate with the HealthConnect* implementation, we will all be beneficiaries of an integrated and interoperable health system, meeting the needs of all participants in a timely manner, and most importantly enhancing health outcomes, and minimising risk."
It's just been a longer journey that I first thought!
Reflecting today, I think that those four words in bold - "grassroots push by consumers" - are important. Health care consumers are now better educated and equipped than ever before to decide what it is that they want from eHealth. Let's complement the top-down national program approach with a powerful, bottom-up, consumer-driven kickstart to eHealth!
More than just requesting our data in a readable format, more than collecting a heap of printed pages or folders of pdfs, we should be requesting our data be in a format that we can do something with, data that we can actually re-use. Think of all the little bits of health information that have been created and left behind after each consultation with your family doctor and at each hospital visit. And of course don't forget your dentist and physio, your naturopath who also prescribes for you nor the doctor you saw only once while on holiday in some far off place. Think of what we could do with all that data; if we could only aggregate all of these snippets of information together. The PHR is the most likely place for our fragmented data to come together; the result being a broader, deeper and richer record than could be achieved by any one provider!
I still firmly believe that Personal Health Records have the potential to transform the way we deliver health care. It may take longer than we all anticipate but the PHR is a very powerful grassroots approach to eHealth. Be patient;-)
*HealthConnect was Australia's eHealth program at the time - now totally MIA and no evidence of it exists online!
Why Is The Shared EHR So Hard?
 'Provision of the right data to the right provider at the right time' is the mantra we commonly hear in eHealth. It sounds deceptively simple! Some promise it; some hope for it; but pretty much no-one has it.
The collective desire for a seamless and efficient shared electronic health record (EHR) which actually provides that data to the provider at a particular time is widespread and yet the EHR remains elusive. If we know what we want, why is the shared EHR so hard to achieve? Why is it taking so long? And what's more, if the financial sector can do it, why not health?
'Provision of the right data to the right provider at the right time' is the mantra we commonly hear in eHealth. It sounds deceptively simple! Some promise it; some hope for it; but pretty much no-one has it.
The collective desire for a seamless and efficient shared electronic health record (EHR) which actually provides that data to the provider at a particular time is widespread and yet the EHR remains elusive. If we know what we want, why is the shared EHR so hard to achieve? Why is it taking so long? And what's more, if the financial sector can do it, why not health?
The progress that we have made in the past 10-20 years of eHealth development has been glacially slow compared to other industries and domains. The approach to health IT, and in particular the shared EHR, has been primarily linear in nature with modest incremental successes achieved. Sure, progress has definitely been made, but despite investing enormous amounts of money and resources, the solution has been more difficult than most ever anticipated. Healthcare doesn't appear to fit the same data interoperability model that has been successful in other domains such as banking or financial services. In a world where connectivity reigns and personal data can flow freely, it is not yet the norm for our health information to be connected nor to flow!
In trying to understand the problem more fully, there are 3 broad headings we need to explore:
- Technical
- Human interaction and activities
- Information exchange
Let's explore some of these issues:
1. Technical
There is little doubt that traditional EHR development has been driven by technology requirements and engineering processes, so many of the typical aspects we consider when thinking of the EHR are actually quite manageable, if not solved - consider storage and retrieval of data, security, role-based access, audit trails, repository storage etc. It is not the technology holding us back here. Some even suggest that the technical aspects of the standalone EHR are the (relatively) easy part, and they may well be right!
With over 7000 EHR vendors in the United States alone, we have proof that building a standalone EHR is undoubtedly achievable. Our EHRs that are in use are traditionally rugged individuals, each created in splendid isolation with the finest data structure, processes and user interfaces! Unfortunately, building proprietary silos of health information is the almost universal approach to EHR development adopted by vendors and does not make it easy for health data to be shared.
I have seen and heard some say that we already have all the standards that we need for eHealth. This is still somewhat controversial and perhaps depends on what part of eHealth that you are trying to make work. For a coordinated eHealth system the desire is for each standards to not only achieve its purpose and goal, but to harmonize with all the others in existence to create a unified whole. Can this vision be achieved? There are certainly some examples of very successful standards. There are also some examples of tweaking of standards for local use... which makes them non-standard again. There are also some examples of standards that have never yet been implemented... what? Let me refer you to the blog post from my colleague - The Crisis in eHealth Standards, by software engineer, Thomas Beale - for an erudite discourse on the strengths and weakness of standards and the standards processes.
2. Human interaction and activities
There is no doubt that some EHR technological achievements have been delayed or even diverted by the human, political and regulatory issues arising from practical implementation - and in most instances, rightly so. We can develop a technological solution, the 'what', but in many jurisdictions the 'how' still has everyone tied up in knots. For example we have technical solutions for unique patient identifiers, data security, and role-based access to data - but 'how' to apply these in practice is more elusive, often crossing into moral and ethical arguments (including confidentiality and privacy), requiring broad social and political agreement and sometimes supportive legislation. These issues have received a disproportionate amount of attention, particularly in the media, and perhaps at the expense of issues that follow which are not so well understood, yet!
Healthcare is not primarily a technical business - it is about people. Yet in our rush towards EHR development we seem to have focused on the EHR being a technical solution rather than merely a tool or medium to support the practical application of health knowledge and provision of healthcare to real people. Some of the most underestimated and misunderstood problems in EHR development are related to:
- the scope and dynamic nature of our health knowledge domain;
- the nature of the human-human interaction that underpins quality health care;
- the approach to capturing and storing the essence of a clinical encounter; and
- the changing clinical requirements, processes and approaches to healthcare provision.
None of these are trivial concepts. They are in the metaphorical 'EHR too hard basket', but we need to address them...
Health is possibly (more likely, probably) the most complex knowledge domain. The extent and scope of health-related knowledge that needs to be represented in an EHR is enormous - it has huge breadth and depth, plus a fine web of complex relationships. Most commonly underestimated is that health knowledge is dynamic - requirements distilled from a clinician and built into an EHR application by a software engineer today can be out-of-date by the time the product is launched.
An encounter between a clinician and a patient is a very complex pas de deux, an intricate communication dance between two parties. Interestingly we clinicians have developed surprisingly effective written methods to capture the information exchange from these encounters, with all the required subtleties and nuances. Capturing the essence of this encounter into a format that can be stored, re-used, queried and shared on a computer is a daunting task and more complex than first appears. Attempting to represent both the data and our clinical processes via the user interface on a computer screen is definitely also an advanced task.
The nature of healthcare provision is also in flux. Consumers/patients/clients/citizens/individuals are increasingly mobile, more now than ever before, demanding healthcare from a range of providers in varying geographical locations. Gone is the old-fashioned notion of the local family physician providing all our needs for all generations of the family; the local physicians are morphing into our health coordinators and facilitators in a world of collaborative and distributed models of care. Our ability to diagnose, treat and prevent conditions are moving with the assistance of new technological advances - in particular, watch out for the potential tsunami-like impact of personalized medicine/genomics on healthcare in the next few years.
3. Information exchange
There are many differing approaches to sharing health information. Why? The plain answer is that it is hard and there is no clear way forward; there are also many differing requirements and starting points:
- Logically, the technical task of sharing health information would be easier if the data was in a common format. Conversely sharing seems to become more difficult by orders of magnitude if our collective data structure is in chaos.
- Practically, some require only the simple exchange of a readable document such as a PDF or semi-structured documents. At the opposite end of the scale, there is need for EHR systems to share detailed and structured health information, so that not only can clinicians and patients read it but the meaning of each piece of data is clearly understood and it can be directly integrated into the computer and utilized in clinical decision-making - this is known as semantic interoperability.
In many places, national eHealth programs and the myriad of 'ruggedly individual' EHR vendors are pursuing technical approaches to data exchange through the set-up of information exchange hubs, message mapping and data transformations. In this instance the focus is on exchange of complete or semi-structured documents as readable health information. Europe and some other parts of the world are taking a different approach, focusing on the exchange of standardized, atomic data and reflected in the European decision to adopt ISO/EN 13606 as its standard for EHR extract exchange.
Exchange of unstructured health-related documents as 'blobs' of data is a great starting point, but in the long term it is a dead end. In reality it is not a sound basis for a shared EHR, nor interoperability, as we can only read them and then store them passively within the EHR. Personally, I want a dynamic, lifelong EHR for myself and my patients - one that can actively create, receive, integrate and re-use my atomic health information and put it to work for me to improve my current health and to provide the basis for future health decisions.
'Provision of the right data to the right provider at the right time' - can this actually be achieved? Clearly the relatively safe, comfortable and incremental technical innovation that has underpinned our EHR development to date hasn't provided the shared EHR solution we hoped for. So at this point let us draw from the wisdom of Einstein: “We can't solve problems by using the same kind of thinking we used when we created them." Indeed, perhaps it is time for a different approach to the shared EHR; one in which we divert our focus and are encouraged to push boundaries; to seek transformational change in our approach to eHealth.
In future posts, I hope to explore some of these issues in more depth and, in turn, some opportunities that arise...
Acknowledgments: Dr Sam Heard and Dr Hugh Leslie

