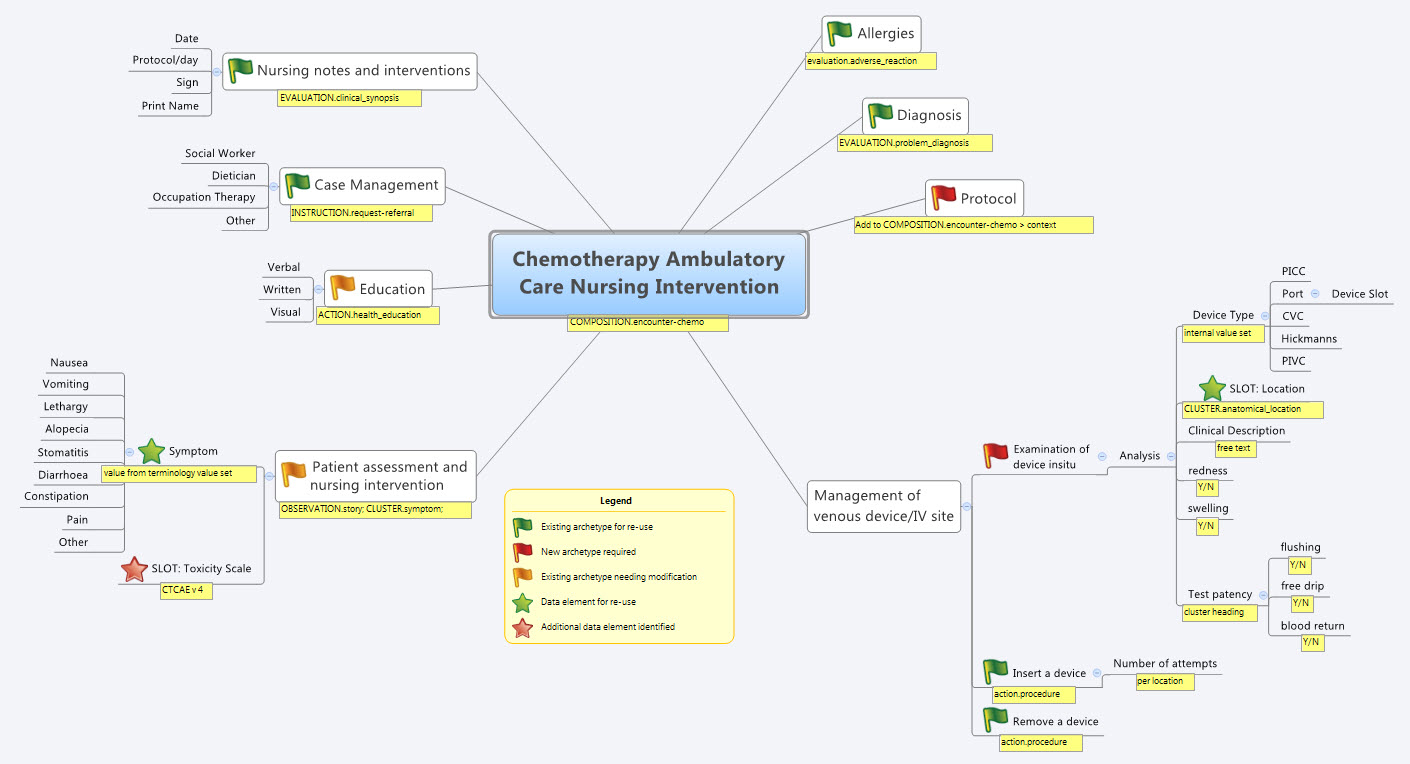
Over the past few days I’ve attempted the task of representing the FHIR Skin and Wound Assessment profiles using openEHR archetypes.
I note that there are three Skin and Wound Assessment FHIR Implementation Guides available online:
"Full CIMI" version – which is the one I chose to model;
I’m a clinical modeler, a clinician by background, so I’m always looking at how we can best represent clinical data in ways that are friendly to grassroots, non-technical clinicians of any sort. The FHIR IG is a tough beast to decipher, despite my experience of gathering patient requirements and turning them into implementable specifications for more than a decade.
My intent as I started this work was specifically that as a clinician I didn’t want to have to fully understand the FHIR representation. I wanted to be able to look at the clinical data and recreate it using my familiar openEHR tooling and representations.
I estimate that it has taken me nearly 2 full days of work – much more than I anticipated - and mostly to trawl through the myriad of online pages for each FHIR resource and associated profile, then to piece together the connections visually so that I could create/reuse appropriate openEHR archetypes and templates. The openEHR representation didn’t take long, largely because of reuse. It was the analysis that was the time killer.
Despite all of that effort, I am still not confident that I’ve got it right. But the following post reflects my experience, plus learnings and some queries.
My modelling assumptions
The three base clinical representations that I’ve gleaned is the Wound Presence Assertion, the Wound Absence Assertion and the Wound Panel Assessment.
Rightly or wrongly, my openEHR templates assume the following:
‘Wound Presence Assertion’ profile is the equivalent of our recording a diagnosis and overview of a wound – so I’ve created a Wound Presence Assertion template, based on the EVALUATION.problem_diagnosis);
‘Wound Absence Assertion’ profile is the positive assertion that a wound is excluded or not present; and
‘Wound Assessment Panel’ is the equivalent of clinical examination findings about a single, identified wound – so I’ve created a Wound Assessment Panel template, based on the OBSERVATION.exam archetype.
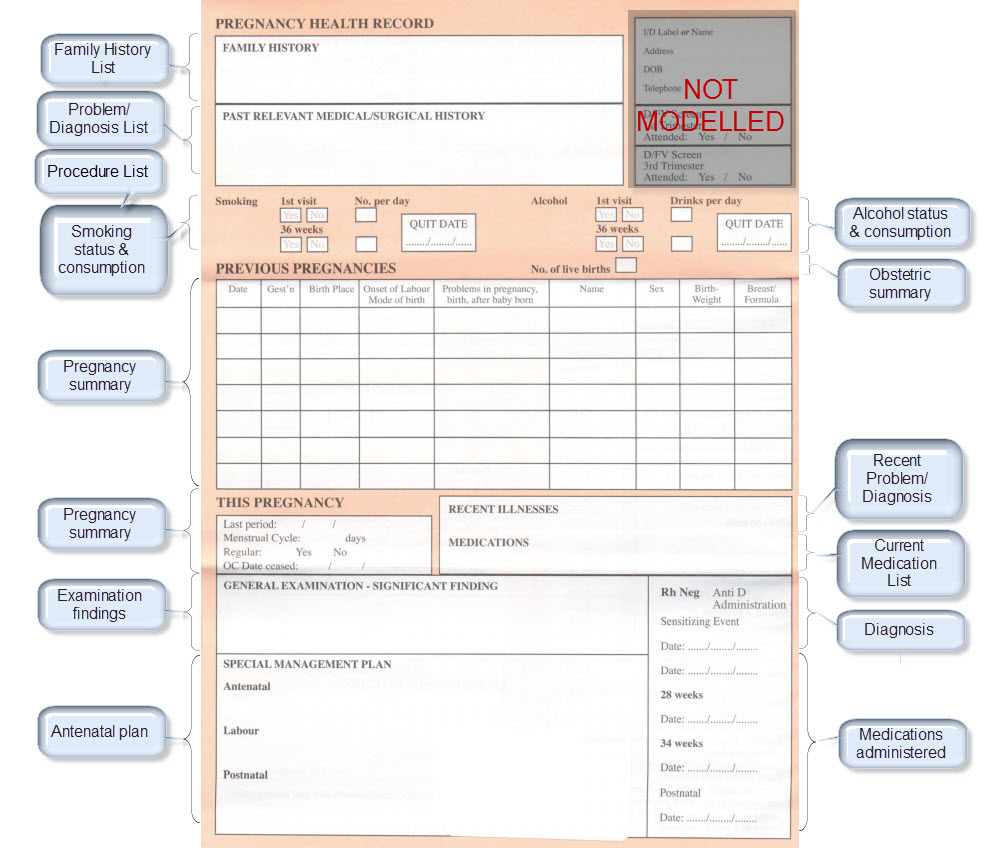
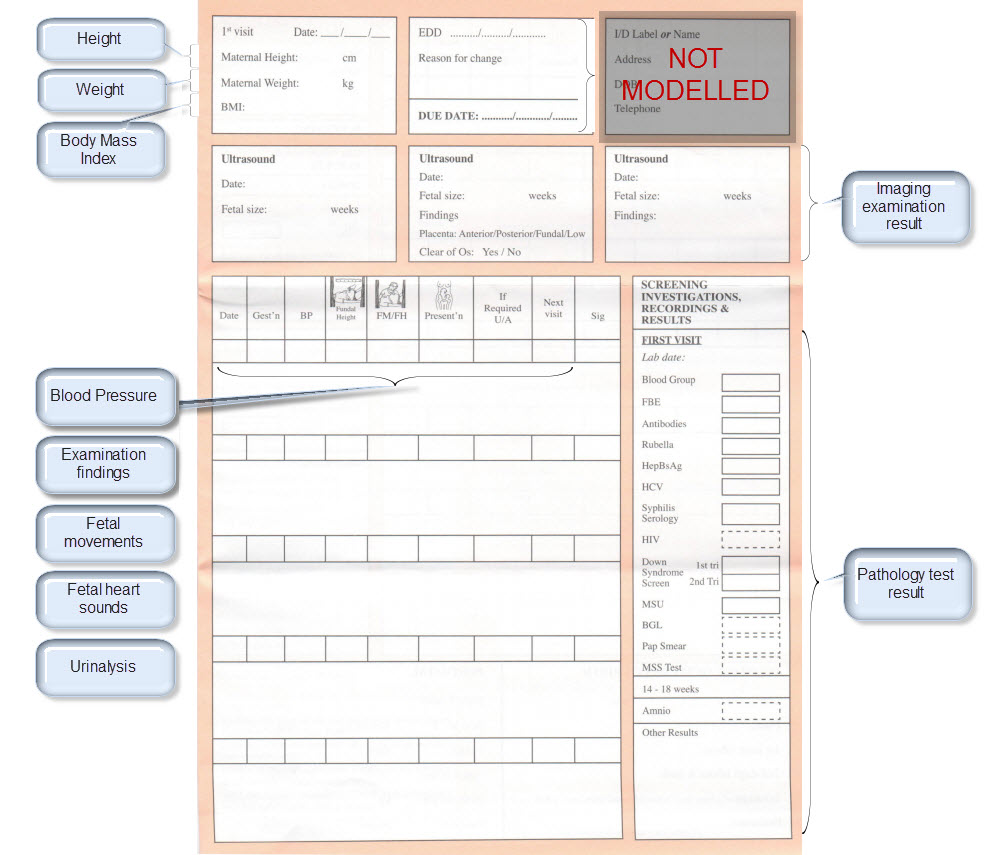
If FHIR components were modelled as 0..1 occurrences then I added them to the archetype at the root level – see the size measurements and edge related data elements in the Examination of a Wound CLUSTER archetype.
If FHIR components were modelled as 0..* occurrences then I added them as repeatable internal cluster groupings – see the Tunnelling and Undermining clusters in the same archetype.
openEHR Representation
You’ll note that I haven’t created a template for the Wound Absence Assertion. I’ll be curious to understand the use case from the FHIR modellers but I cannot understand the use case where a clinician will explicitly record that a wound is absent, not there. They will record that a previously known wound has healed over, or that the skin in the area is normal. But to record that a pressure ulcer on the right buttock is absent – I don’t think so, not even in a medico-legal scenario! If someone can provide me with a use case, I’m happy to reconsider…
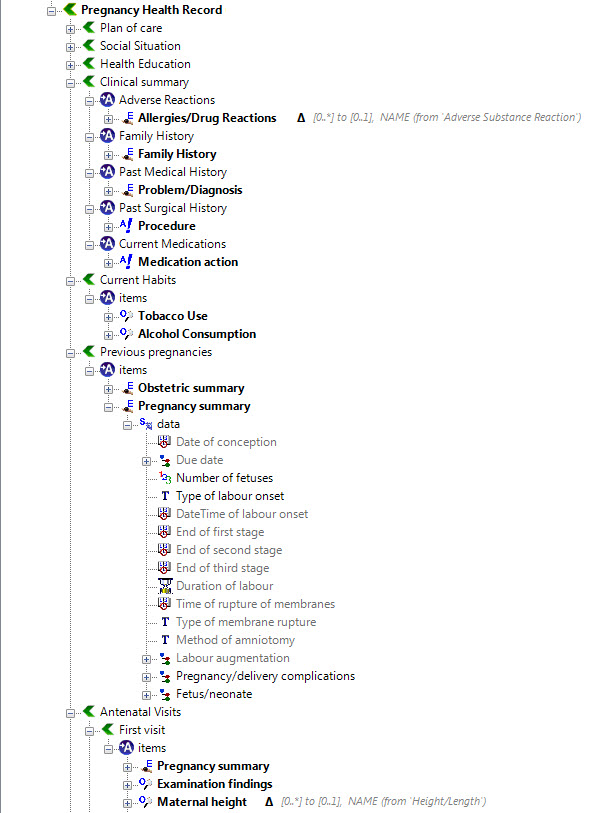
I’ve uploaded the resulting two templates and the associated archetypes to a public incubator on CKM. You can view them all here: https://ckm.openehr.org/ckm/#showProject=1013.30.9.
The two templates comprise 6 reused archetypes, and I created two new archetypes:
Wound assertion details CLUSTER to extend the Problem/diagnosis archetype with FHIR and wound specific content; and
Examination of a Wound CLUSTER representing the examination findings of the wound.
The clinical content within the FHIR IG is generally very sound, and I can see that a lot of work has gone into the development of it and especially the value sets. It is a very useful resource, if you can discern the content in amongst all of the rest of the tech spec. I must admit I got very frustrated and very confused and had to restart a few times.
Once I’d teased it out, I’m very grateful to those who did the hard yards of clinical analysis that underpins this Skin and Wound assessment. Credits on the IG attribute the domain content analysis to Susan Matney from Intermountain Health. I have some other questions for clarifications and would like to discuss a few issues but otherwise this is a really sound piece of work and I’m very pleased with the end result in openEHR.
The templates are up on CKM for you to take a look at:
The rather ugly CKM default UI for templates is deliberately designed for displaying the individual data elements and most of the relevant constraints – much easier for a clinician to be able to review and approve from a content point of view. CKM doesn’t display the URL to value sets, although if you download the .oet and .opt files you will find them safely stored within the code.
Questions, issues
General modelling
I’ve reconfirmed that the openEHR reference model is a godsend. That the data types have set attributes is a given and doesn’t need to be represented over and over again is something I now appreciate enormously, including null flavours etc. Brilliant. There is so much endless repetition in the FHIR resources for RM related data and it takes ages to locate the real clinical data amongst everything else.
Wound Presence Assertion
Anatomical location of the wound is only recorded in this model. I’m not so sure about whether this is a good idea. I think that anatomical location should also be modelled for each examination event (ie included in the Wound Assessment Panel) so that what is being examined is clear and unambiguous. At present the anatomical location of the wound represented by the Assessment Panel appears to only be associated with the Presence Assertion via a common identifier (WoundIdentifier).
Note that I can only find the Anatomical location model in the Logical model and not in the Profile, so maybe I’m missing something?In the logical model, Laterality is represented as ‘Unilateral left’, ‘Unilateral right’ and ‘Bilateral’. I totally agree with the left and right but I have a major problem with identifying one (or more) wound(s) as ‘bilateral’. ‘Bilateral’ should probably not refer to direct observational exam findings at all, but is may have some value in recording conclusions. For example, in examination of each ankle, the finding of pitting oedema may be made, but each side is likely to need explicit recording of different severity or association with ulceration etc, so the findings from each side should be recorded one separately. However, the conclusion of the clinician, at a higher level of abstraction in a physical findings summary or as a diagnosis, may well be bilateral ankle oedema, but it is not advised for use at the point of recording the examination findings.
Given that this representation of anatomical location is in the Assertion and as best I can tell the concept being modelled is a single Wound (SNOMEDCT::416462003), the notion of a bilateral wound is not appropriate.Clinical status values – now these were tricky. We have an existing ‘Problem/diagnosis qualifier’ archetype. It is a messy beastie, largely because clinicians are notoriously messy at how they record these kind of things. The FHIR value set used in this template comprises values from 4 (yes, four) data elements that we have identified as having completely different axes in our archetype. The FHIR value set is drawn from parts of each of our ‘Active/Inactive’, ‘Resolution phase’, ‘Remission status’ and ‘Episodicity’ data elements.
Wound Assessment Panel
As above, I’m concerned that there is no explicit recording of the Anatomical location/laterality parameters so that we can track examination findings over time, especially if there are multiple wounds. An identifier as a connector seems a little flimsy for me.
The concept seems to be a generic wound, but the data elements seem to be focused on recording the findings of an open, ulcerated lesion in reality. If the wound was a long laceration, for example, there are parameters missing such as beginning and end point, relative location to a body landmark. An animal bite might also be difficult to record at the best of times, and something simple like a narrative description would also be helpful.
There is a data point about a pressure ulcer association, with two values – device and pressure site, which reinforces the focus on a pressure ulcer and is very specific. I have modelled that same data point as a repeating cluster pattern of a ‘Factor’ associated present/absent attributes to make this model more applicable to a range of wounds.
Tunnelling is a tricky concept to model. In the FHIR model, tunneling seems to assume that the tunneling radiates out from the edge of the wound but doesn’t allow for deep tunneling from the middle of the wound to be recorded.
The use of a clockface direction is common in a few clinical scenarios, including this one. However the openEHR experience has identified that in order to represent it accurately a few assumed items need to be recorded, such as identification of the central landmark around which the clockface is oriented, as well as the anatomical landmark that identified the 12 o’clock reference point. See our recently published Circular anatomical location archetype.
The Undermining model is represented in the archetype/template as a repeating CLUSTER to allow multiple measurements of the amount undermined in different directions. In the FHIR model, the length and direction are both optional. In the archetype I’ve made the length mandatory as there is no point recording a direction by itself.
I did not model exudate volume in the template as it is measured, and it is not clear to me how you measure a volume at an examination (assuming this assumption about the recording context is correct). Rather it is usually recorded over time, and so does not seem to belong here. I did model the Amount, with a value set that is not available to view, as I assume it is descriptive and could be appropriate here.
Episode is modelled within this context, however it seems to me that it is probably better placed in the Assertion. See the ‘Episodicity’ data element within the Problem/diagnosis qualifier archetype.
Similarly ‘Trend’ feels a little tricky in the context of examination findings. I guess that it could be useful if part of a sequence of exam findings and if it correlated back to the longer timeframe of ‘Course description’ narrative within the Problem/Diagnosis archetype.
There are a few data element which record Yes/No/Unknown answers as though it is answering a questionnaire within the context of recording exam findings. In openEHR we tend to record these as findings that are Present/Absent/Indeterminate so that we can bind, arguably, more meaningful codes to each value and not mix history-like recording with observations.
For a grassroots clinician to review the content there is no doubt that the IG is next to impossible. Perhaps the FHIR community has a more clinician-friendly view that I’m not aware of. It is absolutely needed!
Wouldn’t it be great if FHIR and openEHR communities could collaborate such that this CKM representation could be used to support the FHIR work… but I’ve probably said that a million times… or maybe more. Perhaps one day <sigh>.