Despite some wins, the transition to EHRs has generally been much slower than we anticipated, much harder than we imagined, and it is not hard to argue that interoperability of granular health data remains frustratingly elusive.
"Smart data, smarter healthcare"
Archetypes: health data bridges
What do we want for our health data - silos of information models for different purposes or ones that bridge multiple use cases? From a series of emails shared on the HL7 Patient Care email list in the past few days...
Grahame Grieve (FHIR, HL7):
"Heather, you need to keep in mind the difference between FHIR and clinical models: it's not our business to say not to exchange data that people do have because some user in an edge case might not understand it. We define an exchange standard, not a clinical UI standard..."
and
"Heather, do not lose sight of the difference between a clinical standard for what care/records should be, and FHIR, which is an IT standard for how care records are."
My response:
"...I am concerned about developing another standard that you state clearly is only designed for exchange and not for what care records should be. If we are not designing to try to harmonise data requirements for health information exchange, how clinical care records are and how clinical care records should be, then we are building siloes of data structures again, that will require mappings and transforms ad infinitum. I’d hate to see us end up with a standard for exchange that can’t be implemented for persistence"...
If we end up with models for exchange, models representing current data in systems (whether or not they represent good clinical practice and models that are regarded as the roadmap for good data, then what have we got? Three sets of data models that perpetuate the nightmare of non-interoperability.
Our openEHR archetypes are attempting to bridge all of these. Use them in whatever context you choose - messages, document exchange, EHR persistence, CDS, secondary use, aggregation and analysis, querying etc. The 'secret sauce' is the use of a second layer of modelling - the template, that allows the correct expression of the archetype appropriate for the context of use.
Mappings and transformations are acceptable where we don't have any choice, especially with legacy data, but they open us up to vulnerabilities from errors, misinterpretation and ambiguity, concerns re data integrity and possible overt data loss. Given the choice, lets work towards creating high quality data that can be re-used in multiple contexts safely.
"We have the capability!"
We have had the technology for a purpose-built openEHR-compliant 'plug & play' platform for some time; standalone applications have been built, but just recently it appears that the practical reality of an multi-application platform is also about to happen. "We have the technology. We have the capability..." Stay tuned.
...Reminds me of my 1970's hero, Steve Austin, the Six Million Dollar Man. With apologies to Steve:
"We have the technology. We have the capability to make the world's first universal health platform. openEHR will be that platform. Better than ever before. Robust health data...application independent...semantically interoperable!"
[youtube http://www.youtube.com/watch?v=K7zNY0I5JNI&w=480&h=360]
Preserving health data integrity
How valuable do we really think health data is? How seriously do we take our responsibility to preserve the integrity of our health data? Probably not nearly as much as we should.
Consider the current situation of most clinicians or organisations when purchasing a clinical EHR system. What do they look for? Many possible answers are obvious, but there is one question that I suspect very few are asking. How many consider what data they will be able to export and convert to another format, preserving the current data integrity, at the end of the typical 5-10 year life span of the application? Am I wrong if I suggest it is not many at all?
Despite all the effort that we clinicians put into entering detailed data to create a quality health record we don't seem to often consider the " What next?" scenario. How much and precisely what data will we be able to safely extract, export, transfer or convert into the next, inevitable, clinical system? Ironically, we are simultaneously well aware that clinical systems have a limited technical life span.
Any and all of the health data in a health record is an incredibly valuable asset to the holder, to the patient (if these are not the same entities) and to those downstream with whom we may share it in the future - in terms of $$ invested; manpower resources used to capture, store, classify, update and maintain it; and not least the future value that comes from appropriate and safe clinical decisions being made upon the integrity of existing EHR data.
Yet we don't seem to consider it much... yet. However, as more clinicians are creating increasing amounts of isolated pockets of health data, we should be thinking about it very hard.
Every time we change systems we put our health data at risk - risk of absolute data loss and risk of possible corruption during the conversion. The integrity of health data cannot be guaranteed each time it is ported into a new system because current methods always require some kind of intervention - mapping, transformations, tweaking, 'cleaning', etc. Small errors can creep in with each data manipulation and which over time, can compromise the safety and value of our health data. On principle we know that the data should not be manipulated, but being limited by our traditional approach to siloed EHR applications, we have previously had little choice.
 We need to change our approach and preserve the integrity of our health data at all costs. After all it is the only reason why we record any facts or activity in an electronic health record - so we can use the data for direct patient care; share & exchange the data; aggregate and analyse the data, and use the data as the basis for clinical decision support.
We need to change our approach and preserve the integrity of our health data at all costs. After all it is the only reason why we record any facts or activity in an electronic health record - so we can use the data for direct patient care; share & exchange the data; aggregate and analyse the data, and use the data as the basis for clinical decision support.
We should not be focused on the application alone.
Apps will come and go, but we want our health data to persist - accurate and safe for clinical use - beyond the life span of any one clinical software application.
I've said this before, but it's worth saying many times over:
It's. all. about. the. data.
One of the key benefits of the openEHR paradigm is that the data specifications (the archetypes) are defined independently of any specific clinical system or application; are based on an open EHR architecture specification; and are publicly available in repositories such as the Clinical Knowledge Manager. It means that any data that is captured according to the archetype specification is directly usable by any and all archetype-compliant systems. Plus the data is no longer hard-wired into a proprietary application so that it is orders of magnitude easier to accurately share or transfer health data than it has before.
Clinical system vendors that don't directly embrace the archetype-technology may still 'archetype-aware', and can choose to use the archetype specifications as a means to understand the meaning of existing archetyped data and integrate it appropriately into their systems. Similarly they can map from their non-openEHR systems to the archetype specifications as a standardised method for data export and exchange.
The openEHR paradigm enables potential for archetype-compliant systems to share the same archetyped data repository - along the lines of an Apple platform 'plug & play' approach, with applications being added, removed or updated to suit the needs of the end-users, while the data persists intact. No more data conversions needed.
Adapted from Martin van der Meer, 2009
Now that's good news for our health data.
To HIMSS12... or bust!
This blog, and hopefully some others following, will be about my thinking and considerations as I man an exhibition booth at the huge HIMSS12 conference for the first time next month… Well, we’ve committed. We’re bringing some of the key Ocean offerings all across the ocean to HIMSS12 in Las Vegas next month. If it was just another conference, I wouldn’t be writing about it. But this is a seriously daunting prospect for me. I’ve presented papers, organised workshops, and run conference booths in many places over the years – in Sarajevo, Göteborg, Stockholm, Capetown, Singapore, London, Brisbane, Sydney, Melbourne – but this is sooooooo different!
The equivalent conference here in Australia would gather 600-800 delegates, maybe 40-50 exhibition booths. Most European conferences seem to be a similar size, admittedly these are probably with a more academic emphasis, rather than such a strong commercial bent, which might explain some of the size difference. By comparison, last year’s HIMSS conference had 31,500 attendees and over 1000 exhibition booths – no incorrect zeros here - just mega huge!
I can’t even begin to imagine how one can accommodate so many people in one location. I have never even visited HIMSS before – we are relying heavily on second hand reports. You may start to understand my ‘deer in headlights’ sensation as we plan our first approach to the US market in this way.
Ocean's profile is much higher elsewhere internationally. Our activity in the London-based openEHR Foundation and our products/consulting skills have a reasonable profile in Australia and throughout much of Europe; and awareness is growing in Brazil as the first major region in South America. In many ways the US is the one of the last places for openEHR to make a significant impression – there are some pockets of understanding, but the limited uptake is clearly an orthogonal approach to the major commercial drivers in the US at present, however we are observing that this is slowly changing... hence our decision to run the gauntlet!
openEHR’s key objective is creation of a shareable, lifelong health record - the concept of an application-independent, multilingual, universal health record. The specification is founded upon the the notion of a health record as a collection of actual health information, in contrast to the common idea that a health record is an application-focused EHR or EMR. In the openEHR environment the emphasis is on the capture, storage, exchange and re-use of application-independent data based on shared definitions of clinical content – the archetypes and templates, bound to terminology. In openEHR we call them archetypes; in ISO, similar constructs are referred to as DCMs; and, most recently, there are the new models proposed by the CIMI initiative. It’s still all about the data!
So, we’re planning to showcase two products that have been designed and built to contribute to an openEHR-based health record - the Clinical Knowledge Manager (CKM), as the collective resource for the standardised clinical content, and OceanEHR, which provides the technical and medico-legal foundation for any openEHR-based health record – the EHR repository, health application platform and terminology services. In addition, we’ll be demonstrating Multiprac – an infection control system that uses the openEHR models and is built upon the OceanEHR foundation. So Multiprac is one of the first of a new generation of health record applications which share common clinical content.
This will be interesting experience as neither are probably the sort of product typical attendees will be looking for when visiting the HIMSS exhibition. So therein lies one of our major challenges – how to get in touch with the right market segment… on a budget!
We are seeking to engage with like-minded individuals or organisations who prioritise the health data itself and, in particular, those seeking to use shared and clinically verified definitions of data as a common means to:
- record and exchange health information;
- simplify aggregation of data and comparative analysis; and
- support knowledge-based activities.
These will likely be national health IT programs; jurisdictions; research institutions; secondary users of data; EHR application developers; and of course the clinicians who would like to participate in the archetype development process.
So far I have in my arsenal:
- The usual on-site marketing approach:
- a booth - 13342
- company and product-related material on the HIMSS Online Buyers Guide; and
- marketing material – we have some plans for a simple flyer, with a mildly Australian flavour;
- Leverage our website, of course;
- Developing a Twitter plan for @oceaninfo specifically with activity in my @omowizard account to support it, and anticipating for some support from @openEHR – this will be a new strategy for me;
- And I’m working on development of a vaguely ‘secret weapon’ – well, hopefully my idea will add a little ‘viral’ something to the mix.
So all in all, this will definitely be learning exercise of exponential proportions.
To those of you who have done this before, I’m very keen to receive any insight or advice at this point. What suggestions do you have to assist a small non-US based company with non-mainstream products make an impact at HIMSS?
Why the buzz about CIMI?
With the recent public statement from the Clinical Information Modelling Initiative (CIMI) my cynical heart feels a little flutter of excitement. Maybe, just maybe, we are on the brink of a significant disruption in eHealth. Personally I have found that the concept of standardising clinical content to be compelling and hence my choice to become involved in development of archetypes. During my openEHR journey over the past 5 or so years it has been very interesting to watch the changing attitudes internationally - from curiosity and 'odd one out' through to "well, maybe there's something in this after all".
And now we have the CIMI announcement...
So what has been achieved? What should we celebrate and why?
At worst, we have had a line drawn in the sand: a prominent group of thought leaders in the international health informatics domain have gathered and, through a somewhat feisty process, recognised that a collaborative approach to the development of a single logical clinical content representation (the CIMI core reference model) is a desirable basis for interoperability across formalisms. Despite most of the participants having significant investment and loyalty to their own current methodology and flavor of clinical models, they have cast aside the usual 'not invented here' shackle and identified a common approach to an initial modelling formalism from which other models will be derived or developed. Whether any common clinical content models are eventually built or not, naming of ADL 1.5 and the openEHR constraint model as the initial formalism is a significant recognition of the longstanding work of the openEHR Foundation team - the early specifications emerged nearly 20 years ago.
At its idealistic best, it potentially opens up a new chapter for health informatics, one that deviates from the relatively safe path of incremental innovation that we have followed for so many years - the reliance on messages/documents/hubs to enable us to exchange health information. There is an opportunity to take a divergent path, a potentially transformational innovation, where the focus is on the data itself, and the message/document/EHR becomes more simply just the receptacle or vehicle for the data. It could give us a very real opportunity to store lifelong health information; simplify data exchange (whether by messages or documents), aggregation, querying and analysis; and support knowledge-based activities such as decision support - all because we will (hopefully) have non-proprietary, common, agreed and fully defined models of clinical content and known transformations between each formalism.
Progress during the next few months will be telling. In January 2012, immediately before the next HL7 meeting in San Antonio, the group will gather again to discuss next steps.
There is a very real risk that despite best intentions all of this will fade away to nothing. The list of participating organisations, including high profile standards organisations and national eHealth programs, is a veritable Who's Who of international health IT royalty, so they will all come with their own (organisational and individual) work experience, existing modelling resources, hope, enthusiasm, cynicism, political agendas, bias and alliances. It could be enough to sink the work of this fledgling group.
But many are battle-weary, having been trudging down this eHealth path for a long time - some now gradually realising that the glacial incremental innovation is not delivering the long-term sustainable answers required for creating 21st Century EHRs as they had once hoped. So maybe this could be the trigger to make CIMI fly!
I think that CIMI is a very bright spark on the health IT horizon. Let's hope that with the right management and governance it can be agilely nurtured into a major positive force for change. And in the future, when its governance is mature and processes robust, we can integrate CIMI into the formal standards processes.
Best of luck, CIMI. We're watching!
CIMI - initial public statement
The following public statement has been released by the Clinical Information Modelling Initiative today:
Public releaseThe Clinical Information Modeling Initiative is an international collaboration that is dedicated to providing a common format for detailed specifications for the representation of health information content so that semantically interoperable information may be created and shared in health records, messages and documents. CIMI has been holding meetings in various locations around the world since July, 2011. All funding and resources for these meetings have been provided by the participants. At its most recent meeting in London, 29 November - 1 December 2011, the group agreed on the following principles and approach.
Principles
- CIMI specifications will be freely available to all. The initial use cases will focus on the requirements of organisations involved in providing, funding, monitoring or governing healthcare and to providers of healthcare IT and healthcare IT standards as well as to national eHealth programs, professional organisations, health providers and clinical system developers.
- CIMI is committed to making these specifications available in a number of formats, beginning with the Archetype Definition Language (ADL) from the openEHR Foundation (ISO 13606.2) and the Unified Modeling Language (UML) from the Object Management Group (OMG) with the intent that the users of these specifications can convert them into their local formats.
- CIMI is committed to transparency in its work product and process.
Approach
- ADL 1.5 will be the initial formalism for representing clinical models in the repository.
- CIMI will use the openEHR constraint model (Archetype Object Model:AOM).
- Modifications will be required and will be delivered by CIMI members on a frequent basis.
- A set of UML stereotypes, XMI specifications and transformations will be concurrently developed using UML 2.0 and OCL as the constraint language.
- A Work Plan for how the AOM and target reference models will be maintained and updated will be developed and approved by the end of January 2012.
- Lessons learned from the development and implementation of the HL7 Clinical Statement Pattern and HL7 RIM as well as from the Entry models of 13606, openEHR and the SMART (Substitutable Medical Apps, Reusable Technologies) initiative will inform baseline inputs into this process.
- A plan for establishing a repository to maintain these models will continue to be developed by the group at its meeting in January.
Representatives from the following organizations participated in the construction of this statement of principles and plan:
- B2i Healthcare www.B2international.com
- Cambio Healthcare Systems www.cambio.se
- Canada Health Infoway/Inforoute Santé Canada www.infoway-inforoute.ca
- CDISC www.cdisc.org
- Electronic Record Services www.e-recordservices.eu
- EN 13606 Association www.en13606.org
- GE Healthcare www.gehealthcare.com
- HL7 www.hl7.org
- IHTSDO www.ihtsdo.org
- Intermountain Healthcare www.ihc.com
- JP Systems www.jpsys.com
- Kaiser Permanente www.kp.org
- Mayo Clinic www.mayoclinic.com
- MOH Holdings Singapore www.mohh.com.sg
- National Institutes of Health (USA) www.nih.gov
- NHS Connecting for Health www.connectingforhealth.nhs.uk
- Ocean Informatics www.oceaninformatics.com
- OpenEHR www.openehr.rog
- Results4Care www.results4care.nl
- SMART www.smartplatforms.org
- South Korea Yonsei University www.yonsei.ac.kr/eng
- Tolven www.tolven.org
- Veterans Health Administration (USA) www.va.gov/health
Further Information:
In the future CIMI will provide information publicly on the Internet. For immediate further information, contact Stan Huff (stan.huff@imail.org)
The DCM environment - a crowdsourcing initiative
There are a number of detailed clinical models available to choose from, from diverse backgrounds, for many differing purposes and with varying degrees of success and penetration. More are arriving on the scene as time goes on – only this week the Clinical Information Models from ONC in the US have been released for their first 2 week period of public comment, and I'd never heard of them before. I haven’t been able to find a single document pointing to an overview on available models – current or past. So, given the current activity development of a quality standard around detailed clinical models in ISO TC 215 and the number of models under development, I thought it timely to try to co-ordinate sourcing a picture of the detailed clinical model landscape.
Well aware that my understanding is limited to the openEHR archetypes I work with, I thought I’d take this opportunity to draw together some of my research and request some assistance from others to crowdsource a picture of the detailed clinical model environment that we operate within.
I have published my initial attempt as a Google doc, see below. The existing information needs to be verified; there are clearly many gaps that need to be filled in; and there are probably other DCMs to be added.
EDIT the DCM environment document...
Anyone can participate, and if you edit, please add your name/org/email to the Contributor list, so that attribution can be made.
Please feel free to edit; to add new DCMs; to correct and verify the facts. Add a column, suggest changes to structure where you see a need... Just make a comment, if you like.
You can clearly see all the gaps in our collective knowledge at present…
Let’s see if we can actually pull together a useful resource. The intent is collaboration to achieve a goal that is possibly greater than most of us can achieve by ourselves.
All contributions are gratefully received and can be freely shared with anyone who is interested.
The latest crowdsourced version of the document appears below. You will need to scroll to seen the entire content.
[googleapps domain="docs" dir="document/pub" query="id=11xGZs4drfwb7TY15Iwpk52UIxZcFhqMcMp1i3yV4To8&embedded=true" /]
________________________________________
Further Comment: 11 August 2011
I've been asked where this work is going and what is planned...
The answer is that I haven’t a specific plan.
To my knowledge there has been no environmental scan to explore what the range of models is, to my knowledge, so thought I’d try myself. I spent a day to develop the initial framework, sparse though it was. Googling was a very frustrating experience and finding out the information that we’re interested in from available websites was not easy.
Hence I thought I’d experiment with the crowd sourcing approach. I know there is a rising tide of interest in the generic concept of detailed clinical models and a lot of confusion about the work in ISO and HL7.
I hoped it might contribute to clarifying the situation and providing a concrete anchor on which to base discussions on detailed clinical models.
Worst case scenario, the final result may be little more than a brainstorming, however it will at least form the basis for anyone interested to launch a more rigorous research program, including potentially feeding into the ISO work. Best case scenario, we may have a significant resource. Final outcome will probably be somewhere in between.
I’m happy to leave it online as a general resource – it seems to have generated a lot of interest. I'm hopeful that it will keep evolving.
Clinical modeling: academic vs practical
In a comment on the previous DCM post, Gordon Tomes sent a link to an interesting 2009 academic paper by Scheuermann, Ceusters & Smith - Toward an Ontological Treatment of Disease and Diagnosis [PDF] There is a place for this 'pure', academic approach as a means to seek clarity in definitions, but a telling sentence in the paper for me is: "Thus we do not claim that ‘disease’ as here defined denotes what clinician in every case refer to when they use the term ‘disease’. Rather, our definitions are designed to make clear that such clinical use is often ambiguous."
This is totally right, but despite the apparent ambiguity this clinicians still manage :)
The approach for archetypes/DCMs is not to get tied up in these definitional knots unnecessarily but to concentrate on consensus building around the structure of the data. Use of ontologies and definitions are key elements in designing and modelling archetypes but the resulting models should not be based on academic theory but on existing clinical practice and processes. Our EHRs need to represent what clinicians actually need to do in order to provide care. Sometimes we need to be practical and pragmatic. For example, I once challenged a dissenter to build a Blood Pressure archetype based purely using SNOMED codes - the result was a nonsense, a model that he agreed was not useable by clinicians.
The approach to building the Problem/Diagnosis archetype has not been to try to differentiate these two terms pedantically. Many have tried to separate a 'Problem' from a 'Diagnosis' for years with little success - no point waiting for this to be resolved, it probably won't. And even if we did make a theoretical decision on a definition for each, the clinicians would still likely classify the way they always did, not the way the academics or standards-makers would like them to do it!
In the collaborative CKM reviews for the NEHTA Problem/Diagnosis archetype we have been able to observe that clinicians and other stakeholders have achieved some consensus around the structured data required to represent the concepts of a problem plus the addition of some extra optional data elements to represent formal diagnoses. After completing only four review rounds, the discussion now is focused on finessing the metadata and descriptions, not on debating the structure of the model.
Clinicians may still go round in circles arguing about what constitutes an actual problem vs a diagnosis - for example, some may classify heartburn as a problem, others as diagnosis. No matter. By using a single model to represent both we can ensure that no matter how heartburn may be labelled by any individual clinician, we can easily query and find the data in a health record, and in addition there is a single common model to be referenced by clinical decision support engines.
A small success, maybe.
openEHR: interoperability or systems?
Thomas Beale (CTO of Ocean Informatics and chair of the Architecture Review Board of the openEHR Foundation) posted these two paragraphs as part of the background for his recent Woland's Cat post - The Null Flavour debate - part I. It is an important statement that I don't want to get lost amongst other discussion, so I've reposted it here:
An initial comment I will make is that there is a notion that openEHR is ‘about defining systems’ whereas HL7 ‘is about interoperability’. This is incorrect. openEHR is primarily about solving the information interoperability problem in health, and it addresses all information, regardless of whether it is inside a system or in a message. (It does define some reference model semantics specific to the notion of ‘storing information’, mainly around versioning and auditing, but this has nothing to do with the main interoperability emphasis.)
To see that openEHR is about generalised interoperability, all that is needed is to consider a lab archetype such as Lipid studies in the Clinical Knowledge Manager. This archetype defines a possible structure of a Lipids lab test result, in terms of basic information model primitives (Entry, History, Cluster, Element etc). In the openEHR approach, we use this same model as the formal definition of this kind of information is in a message travelling ‘between systems’, in a database or on the screen within a ‘system’. This is one of the great benefits of openEHR: messages are not done differently from everything else. Neither is interoperability of data in messages between systems different from that of data between applications or other parts of a ‘system’.
Engaging Clinicians in Clinical Content (in Sarajevo)
Just browsing and found the link to our presentation for a paper the CKM team gave at the Medical Informatics Europe conference in Bosnia, 2009. Thought I'd share it here, in memory of an amazing conference and location:
MIE09: Engaging Clinicians in Clinical Content [PDF]
Our presentation:
[slideshare id=1958920&doc=engagingcliniciansinclinicalcontent-090906091020-phpapp02]
And the reference to Herding Cats is explained (a little) in the embedded video - actually an advertisement for EDS:
[youtube http://www.youtube.com/watch?v=Pk7yqlTMvp8&w=425&h=349]
I arrived after a 37 hour flight - Melbourne - London - Vienna - Sarajevo to join my Ocean CKM team - Ian McNicoll and Sebastian Garde. We presented our paper and also ran an openEHR workshop.
The conference was held at the Holiday Inn in Sarajevo - the hotel in which all the international journalists were holed up during the war, on 'Sniper Alley'.
Despite it being 14 years after the war had ended, raw emotions were palpable many times during the conference. That a pan-European conference was being held in his beloved Sarajevo under his auspice was a very overwhelming event for the Professor of Informatics and he was often seen in tears!
From my hotel room window in the centre of the city I could see 6 cemeteries.
Walking through the inner city we came across many cemeteries, thousands of marble gravestones with the majority representing the young men aged between 18 & 26.
Bullet holes were still very obvious on the walls of buildings.
Yet the city was vibrant, alive and welcoming.
I won't ever forget this visit.
Some photos of the experience:
Anatomy of a Problem... a Diagnosis...
Anatomy of an Adverse Reaction
 Discussion about how to represent some of the commonest clinical concepts in an electronic health record or message has been raging for years. There has been no clear consensus. One of those tricky ones - so ubiquitous that everyone wants to have an opinion - is how to create a computable specification for an adverse reaction.
In the past few years I have been involved in much research and many discussions with colleagues around the world about how to represent an adverse reaction in a detailed clinical model. I'd like to share some of these learnings...
Discussion about how to represent some of the commonest clinical concepts in an electronic health record or message has been raging for years. There has been no clear consensus. One of those tricky ones - so ubiquitous that everyone wants to have an opinion - is how to create a computable specification for an adverse reaction.
In the past few years I have been involved in much research and many discussions with colleagues around the world about how to represent an adverse reaction in a detailed clinical model. I'd like to share some of these learnings...
Making sense of 'Severity'
 'Severity' is a pretty simple clinical concept, isn't it?
I thought so too until I first sat down to create a single, re-usable archetype to represent 'Severity'. I soon discovered that I had significantly underestimated it - the challenge was greater than appeared at first glance.
'Severity' is a pretty simple clinical concept, isn't it?
I thought so too until I first sat down to create a single, re-usable archetype to represent 'Severity'. I soon discovered that I had significantly underestimated it - the challenge was greater than appeared at first glance.
The archetype development process is usually relatively straight forward - a brain dump of all related information into a mind map, followed by rearranging the mind map until sensible patterns emerge. They usually do, but not this time.
However, the more I investigated and researched, the more I could see that clinicians express severity in a variety of ways, sometimes mixing and 'munging' various concepts together in such a way that humans can easily make sense of it, but it is much harder to represent in a way that is both clinically sensible and computable.
The simple trio of 'Mild', 'Moderate' and 'Severe' is commonly used as a gradation to express the severity of a condition, injury or adverse event. Occasionally these are defined for a given condition or injury, however most often these are subjective and there should be concern that one clinician's mild might be another's moderate. Some also include intermediate terms, such as 'Mild-to-moderate' and 'Moderate-to-severe', but one has to begin to be concerned if these more subtle differences make the judgement even more unreliable, especially if we need to exchange information between systems.
Others add 'Trivial' – is this less than 'Mild'? By how much? Is there a true gradation here? And what of 'Extreme'? Still others add 'Life-threatening' and 'Death' – but are these really expressing severity? Or are they expressing clinical impact or even an outcome?
When exploring severity I found examples of severity expressed by clinicians in many ways. This list is by no means exhaustive:
- Pain – expressed as intensity, often including a visual analogue scale as a means to record it ie rate from 0-10
- Burns – describing percentage of body involved e.g. >80% burns
- Burns – describing the depth of burn by degree e.g. first degree
- Perineal tears – describe the extent and damage by degree – e.g. second degree tear
- Facial tic – expressed in using frequency e.g. occasional through to unrelenting
- Cancer – expressed as a grade, clinical or pathological
- Rash – extent or percentage of a limb or torso covered.
- Minor or major
- Mild, disabling life-threatening
- Relative severity – better, worse etc
- Functional impact – ordinary activity, slight limitation, marked limitation, inability to perform any physical activity
- Short or long term persistence
- Acute or chronic
In practice, clinicians can work interchangeably with any of these expressions and make reasonable clinical sense of it. The challenge when creating archetypes, and other computable models, is to ensure that clinicians can express what they need in a health record, exchange that information safely with other clinicians, allow for knowledge-based activities such as decision support.
In addition, there also needs to be a clear distinction between 'severity' and other, related qualifiers that provide additional context about 'seriousness' of the condition, injury or adverse reaction. These sometimes get thrown into the mix as well, including:
- Clinical impact (or significance);
- Clinical treatment required; and
- Outcomes.
Clinical impact/significance might include:
- None - No clinical effect observed
- Insignificant - Little noticeable clinical effect observed.
- Significant - Obvious clinical effect observed
- Life-threatening - Life-threatening effect observed
- Death – Individual died.
Immediate clinical treatment required might include:
- No treatment
- Required clinician consultation
- Required hospitalisation
- Required Intensive Care Unit
Outcomes might include:
- Recovered/resolved
- Recovering/resolving
- Not recovered/resolved
-
Fatal
Then there are those that recovered or the condition resolved but there were other sequelae such as congenital anomalies in an unborn child...
Severity ain't simple.
Over time, and repeatedly returning to it when modelling it in various archetypes including Adverse Reaction and Problem/Diagnosis, I've come to the conclusion that I don't think it is useful or helpful for us to model 'severity' in a single, re-usable archetype. It seems to work better as a single qualifier element within archetypes – then we can bind it in to terminology value sets that are useful for that specific archetype concept and for the use-case context.
When a clinical knowledge pattern is easily identified, creating an archetype is easy - the archetype almost writes itself! So over time I've learned not to try to force the modelling. Some archetypes 'work'; some, like this one, just don't.
Anatomy of an archetype
 With this blog I want to establish a simple baseline statement or overview about openEHR archetypes, aggregated from other posts and publications – a reference point if you like – from which we can journey further and in more detail into the issues around clinical modelling using specific archetypes.
Archetypes are a strong basis for data liquidity. Create the archetype once, agree through peer-review, re-use when and where required – the foundation for a 'universal health record'.
With this blog I want to establish a simple baseline statement or overview about openEHR archetypes, aggregated from other posts and publications – a reference point if you like – from which we can journey further and in more detail into the issues around clinical modelling using specific archetypes.
Archetypes are a strong basis for data liquidity. Create the archetype once, agree through peer-review, re-use when and where required – the foundation for a 'universal health record'.
Definition:
Formal Definition
An archetype is a computable expression of a domain content model in the form of structured constraint statements, based on a reference (information) model. openEHR archetypes are based on the openEHR reference model. Archetypes are all expressed in the same formalism. In general, they are defined for wide re-use, however, they can be specialized to include local particularities. They can accommodate any number of natural languages and terminologies. [Archetype Definitions and Principles]
Clinician's 'lay' definition:
An archetype is a structured, computable specification for one single, discrete clinical concept. - much simpler!
Each archetype is a rich health data specification, defined and agreed by the clinicians themselves to ensure that each model is 'fit for clinical purpose'. Collectively these archetypes together can create an electronic health record 'lingua franca', or an example of PCAST report's 'Universal Exchange Language (although archetypes are not limited only to exchange of health information).
Archetypes can be used to model a range of clinical concepts:
- Simple and straightforward - such as 'Blood Pressure'; 'Body Weight'; or 'Symptom'; or
- Complex, compound concepts - such as 'Medication order', or recording how a 'Procedure' has been carried out.
Purpose
Archetypes are:
- Potential 'agents for change' – allowing knowledge-driven EHRs as an orthogonal approach to EHR development.
-
The basis for a coherent and consistent approach for the whole continuum of clinical care and related activities:
- Recording, storing and querying health information in electronic health information;
- Exchanging health information - the broader the agreement & governance, the greater the potential for interoperability;
- Data aggregation;
- Knowledge-based activities; and
- Comparative data analysis.
Using archetypes requires:
- A change of mindset – no longer silos of data; no longer message and document driven.
- Upfront planning & coordination in development of archetypes, but not necessarily more time
-
CLINICIAN LEADERSHIP & ENGAGEMENT
- To ensure the quality of clinical data in our EHRs
- To warrant the clinical data is safe & 'fit for purpose'
Design
Each archetype is designed:
- As a maximal data set effectively everything one can think of about a clinical concept in all situations by the consumer and any and all providers. This design intent is pragmatically constrained to be sensible in some situations, however good archetype design will always ensure that the full model can be achieved through nesting of additional archetypes to create the maximal dataset/universal use-case ideal.
- For the 'universal use-case' – for re-use in any and all scenarios, from use at home, to primary care, community care, hospital care, secondary use and for research.
Each archetype needs to represent the variety of data that is required to represent the information required for all aspects of clinical care and related activities, including:
- The ability to capture both free-text vs structured data;
- Normal statements such as "Nil significant" or "NAD";
- Graphable data;
- Images – eg a homunculus, or a surgical diagram;
- Multimedia including photos, video and audio or an ECG wave format;
- Questionnaires, checklists etc
Think about how the clinician may record the data: different clinicians will need differing approaches to recording their health information and the models need to allow for this clinical diversity. Some will prefer to use more free text and others more structure; some need more detail than others; we need to capture current requirements for recording and exchange while keeping in mind what might be best practice in the near and distant future.
The best designers, without a doubt, have a clinical background. I have seen clinicians and non-clinicians given identical clinical content, but the archetypes produces are surprisingly different. Clinician modellers definitely model their domain better than those who are not familiar with clinical practice. They are perhaps better able to envision the fractal nature of medicine and take that in to account in creating archetypes, especially the complex areas which require nesting of archetypes within each other to capture the depth and richness required in the models.
Integration with terminologies such as SNOMED, ICD or LOINC are strategic and important – the structure of an archetype is not enough to represent clinical information adequately, and similarly terminology alone is inadequate. However the combination of terminology within an archetype structure, by naming elements or provision of value sets, is semantically very powerful. There is still considerable work to be done to determine how to represent the information overlap – that which can be expressed either in the archetype structure itself or via terminology. This remains a 'grey zone' and is a pressing area for collaboration or research.
Archetype classes – support clinician's processes
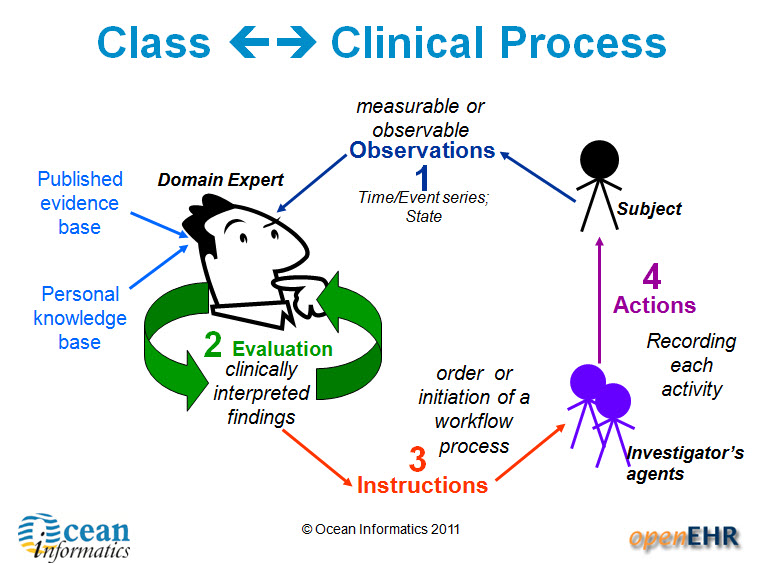
There are four main categories of archetypes that are useful to understand – each corresponding to classes in the openEHR Reference Model.
1. Compositions – which correspond to commonly used clinical documents, such as 'antenatal visit' or 'care plan'. 2. Sections – these are effectively used to assist with human navigation within EHRs and correspond to document headings, for example 'antenatal examination' or 'summary'.
3. Entries – these are the most common and are fundamental building blocks of EHRs. There are four main types of Entry archetypes:
- Observations – recording measurable or observable data e.g. blood pressure, symptoms or weight;
- Evaluations – recording clinically interpreted findings e.g. adverse event, diagnosis or assessment of risk;
- Instructions – recording the initiation of a workflow process, such as a medication order or referral;
- Actions – recording clinical activities e.g. procedure or medication administration. Actions complement the instruction and can record the ensuing state of the instruction, such as 'completed' or 'cancelled'.
4. Clusters
Lifecycle & Governance
The Clinical Knowledge Manager (CKM) – www.openEHR.org/knowledge - is an international, online clinical knowledge repository under the auspice of the openEHR Foundation that provides:
- Access to a library of cohesive archetypes and related knowledge artefacts;
- Peer review, life cycle management & publication process via a Web 2.0 approach for clinical content, terminology binding, terminology reference set binding and translations;
-
Clinical Knowledge governance underpinned by a digital asset management system and providing:
- Provenance, audit trails, validation checking
- Release sets for implementation
Archetypes are freely available under a Creative Commons license.
Use
The need for EHRs to be able to represent clinical diversity is absolutely underrated. Existing EHR systems corral clinicians into certain types of recording etc, but for safety we need more diverse ways for clinicians to record what they need for patient care, and also to ensure that what is exchanged is appropriate for the patient and their individual circumstances. A 'one size fits all' document approach to emergency summaries or messages can be potentially unsafe. Elderly, pregnancy, chronic disease, paediatrics are extremely common examples where there are specific additional requirements needed to summary data sets
Templates are computable specifications for a specific clinical scenario or use-case. The openEHR paradigm takes the governed and agreed archetypes from CKM and creates clinically useful specifications that can be implemented in systems by:
- Aggregating the necessary archetypes together; and
- Constraining the maximal dataset archetypes so that only relevant data elements are active.
Examples include all clinical content required for a specific Message, Document, Clinical consultation or Report e.g. a histopathology report, a referral, an antenatal visit or a discharge summary.
In this way we can, in colloquial terms, 'have our cake and eat it too'. At the same time, CKM ensures tight clinical knowledge governance, yet templates allow for expression of clinician diversity.
Benefits
Archetypes provide a 'glide path' to knowledge-level interoperability of health information. They can:
- Bootstrap new application development – a source of agreed clinical content, avoid re-inventing the wheel for each vendor, organisation or project;
- Provide a forward 'roadmap' for existing applications – supporting gradual transition from proprietary silos to common data representations, perhaps via mapping to common messages as an interim step;
- Support data integration – the means to migrate valuable silos of legacy data to a common, re-usable representation
- Enable data aggregation & comparative analysis – the basis for ensuring 'apples' are 'apples', not 'oranges';
- Simplify messaging – keep message content consistent with EHR content;
- Enable knowledge-based activities eg Clinical Decision Support Systems – consistent & coherent; create once, re-use in all systems.
Power drills, Cardiologists and collaborative consumption
This week I watched Rachel Botsman's TEDxSydney talk: Collaborative Consumption. Rachel's premise is that we're "wired to share", and I particularly liked her illustration at the 10 minute, 30 second mark... ASSUMPTION #1: Most people own a power drill.
ASSUMPTION #2: Most power drills are used for a total of 12-13 minutes in their entire lifetime << seems not unreasonable.
NEED: We actually need a hole, not a drill!
CONCLUSION: Either rent a drill from someone else, or rent yours to everybody else.
WHAT IS THE END GOAL?: Share the resources better!
[ted id=1037]
While visiting Brazil last year , I learned that in São Paulo if you get referred to a Cardiologist, you don't get to choose which one, you just get sent to one. That doesn't sit well with me, especially as a clinician, and I'm pretty careful to whom I entrust my care, or my family's care.
Yet apparently the waiting time is down to trivial time frames. People are actually getting treated. That is significant!
Apparently resources are being used better, simply because of a central booking system and an algorithm matching clinician with patient and location.
When you start to add in some of the benefits from eHealth such as potential shared EHRs, this starts to make more sense. I will still struggle with the lack of choice, but if patients are being seen more efficiently... then maybe it is a good, or even better, thing.
Stop for a moment and consider:
If we want health reform...
If we want more efficient use of resources for our health $$$...
... then we need to think of how to better use Health IT to support the notion of collaborative health consumption - both of resources & information. How can we better match available resources with need? How can we enable consumer choice at the same time?
There's a tension there - I can foresee many issues, but also opportunities.
I have more questions than answers.
Very interested in your opinion.
Don’t re-invent the (clinical content) wheel...
 It was with great interest that I read about the the recommendation for a universal exchange language in the recent release of the US report to the President: REALIZING THE FULL POTENTIAL OF HEALTH INFORMATION TECHNOLOGY TO IMPROVE HEALTHCARE FOR AMERICANS: THE PATH FORWARD.
I had asked the Direct project about the existence of a national plan for standardising clinical content only recently... It appeared that here was a plan after all.
It was with great interest that I read about the the recommendation for a universal exchange language in the recent release of the US report to the President: REALIZING THE FULL POTENTIAL OF HEALTH INFORMATION TECHNOLOGY TO IMPROVE HEALTHCARE FOR AMERICANS: THE PATH FORWARD.
I had asked the Direct project about the existence of a national plan for standardising clinical content only recently... It appeared that here was a plan after all.
So, to the report. The approach and benefits proposed started well...
The best way to achieve a national health IT ecosystem is to ensure that all electronic health systems can exchange data in a universal exchange language. The systems themselves could be designed in any manner desired — they could accommodate legacy systems that prevail or new recordkeeping systems and formats. The only requirement would be that the systems be able to send and receive data in the universal exchange language. (p41)
I have previously blogged about a universal health record underpinned by an application independent library of clinical content definitions, so the intent and benefits are well aligned with my preferred approach.
But then alarm bells started to ring....
Because of its multiple advantages, we advocate a universal exchange mechanism for health IT that is based on tagged data elements in an extensible markup language. If there were another equally good solution, it should also be considered; we have collectively been unable to think of one. (p43)
Issue #1: Isn't it more appropriate for step one to identify the need for standardised clinical content as a policy, rather than specify the format up front? Isn't that really the domain of health informatics experts as part of a subsequent work plan? I feel like we've skipped a couple of steps in the decision-making process. And are they really advocating the creation of this metadata-tagged XML from a zero starting point?
Issue #2: The last 9 words of that paragraph, "...we have collectively been unable to think of one." I'm glad that they are still open to equally good solutions being considered as indeed there are many ways that individuals, groups and organisations are exploring how to standardise clinical content definitions as the basis for a universal exchange mechanism.
In ISO TC 215, the International Standards Organisations Technical Committee for Health Informatics, there is a new work item which has been evolving for at least 2 years, although yet to attain committee draft status, known as ISO 13972 - Quality criteria for detailed clinical models. This work item is targeting a new international standard for determining quality criteria about the development of detailed clinical models - all clinical models, pick your flavour! In the world of international standards it has been recognised for years that with the plethora of different approaches to developing clinical models for EHRs, there is a need for some criteria to support quality aspect in their development. This work is being led by modellers from the Netherlands, with experts participating from the Australian, Danish, German, Swedish, US and Canadian standards organisations. Creating clinical content is definitely not a new field of endeavour by the time it enters the international standards arena.
So, I am extremely surprised that this expert PCAST group have not been able to 'collectively think' of an existing alternative.
In my last blog - Clinical Knowledge Governance in a Web 2.0 world – I pointed to a number of approaches to standardised clinical content to support health information exchange.
1. In the US – including, but by no means limited to:
- the HL7 standards organisation - where my UK colleague, Charlie McKay, informs me that there are more than 20 different approaches to clinical content development. Keith Boone (@motorcycle_guy) has posted his response to the PCAST report from a HL7 point of view - The Language of HealthIT;
- Stan Huff's group at Intermountain Health in Utah have had extensive experience in defining standardised clinical content across all of Intermountain's systems – they are leading experts in this domain; and
- I understand Don Mon and his team from AHIMA have also been working in this area.
2. In Europe, and Australia:
- The Australian National eHealth Transition Authority (NEHTA) has just launched a national clinician-led CKM approach to grassroots clinical content standardisation creating NEHTA's Detailed Clinical Models (DCMs).
- The Swedish national eHealth program has been actively working to integrate openEHR archetypes with its' VTIM architecture.
In addition, a few more points...
Firstly, the focus of the PCAST report is still only on data exchange, not on ensuring a sound foundation of a person-centric electronic health record. I'll say it again... get the data right and then the data will be able to be re-used, to multitask, be liquid, flowing to where it needs to be. It will become the solid foundation on which to build lifelong health records, simpler health information exchange, data integration & aggregation, research, reporting and knowledge-based activities. By focusing on exchange alone, then... you'll hopefully be able to exchange well and the rest will be considerably more uncertain.
Secondly, the proposed variant of XML is described as a 'straightforward' and 'superior' solution (p44), and the assumption that it will be scalable, protected by encryption, and that data element access services will be enough to support the health information exchange required. By contrast HL7, ISO/CEN 13606 and openEHR have taken decades to develop and refine underlying reference models to ensure that they have an unambiguous, consistent, secure way to represent personal health information – so you know who created the data, who is the subject of care, what the data means, what are the access rules applicable etc. In the openEHR environment, the specification authors developed Archetype Definition Language (ADL) for the purpose - and now part of the ISO 13606 standard - because the alternatives such as standard XML were not robust enough to represent health information. A 'straightforward' XML approach has a strong possibility of failure without a RM underpinning it.
And finally, there is the area of clinical knowledge governance itself. Health is dynamic, complex and diverse. The work required to represent healthcare as computable clinical content definitions or specifications is huge – don't underestimate the sheer volume of work that will be required. It is not realistic to expect a 'rapid mapping' of existing proprietary data structures into tagged data elements. Who will decide the clinical content in the models? If there are over 7000 clinical vendors in the US, which will be 'the source' or sources? Which are 'correct' or 'authoritative'? What methodology will be used to create the models? What level of granularity for each clinical element? How will they be aggregated together to represent clinical documents or events, and constrained to be useful for the clinical purpose? I have a million more questions...
Once the information models are defined, there will be a need for them to be validated before they can become the basis for a standardised or national clinical content library – suitable for consumers, clinicians, organisations, vendors, researchers and jurisdictions. A requirement will be recognised for life-cycle management and publication of these models, roadmaps for legacy data to migrate towards, and harmonise with, the new national health information 'source of truth', plus ongoing maintenance and governance.
Eric Browne stated in his recent blog, Recasting e-Health in the USA:
The work in Sweden, the UK, Singapore and even Australia, based on openEHR or ISO 13606 archetypes (i.e. implementable renditions of Detailed Clinical Models) is far more advanced and promising than that offered by the PCAST approach.
openEHR, which is my interest, has an approach to defining, agreeing and governing clinical content models for electronic health records, known as archetypes. It has taken more than 18 years to develop the openEHR technical specifications, and the last 10 years to achieve its' current approach and position in terms of clinical modelling. It is gaining traction, albeit with a modest volunteer community, especially now that it has a collaborative portal, known as the Clinical Knowledge Manager, to support sharing or models, reviews of clinical content, translation and terminology binding, and model governance.
Standardising health information definitions for health records or exchange is not a trivial task. Learn from what has already been achieved – all shapes, flavours and doctrines. Whatever you do, don't reinvent the wheel and create yet another universal language!
Can clinicians agree?
 There have been a number of robust discussions in recent weeks around the claim that clinicians achieve consensus around computable definitions for clinical content. All discussions have been with MDs, some with substantial experience in various international standards organisations. All have been extremely sceptical...
There have been a number of robust discussions in recent weeks around the claim that clinicians achieve consensus around computable definitions for clinical content. All discussions have been with MDs, some with substantial experience in various international standards organisations. All have been extremely sceptical...
The first challenge: "What is a heart attack?” - and the response, “5 clinicians, 6 answers” – and they are probably very accurate!
The second: “What is an issue vs problem vs diagnosis?” - I'm told that this has been an unresolved issue in HL7 for 5 years+!
And the third, from an obstetrician: “The midwives want all this rubbish that we don’t” - perhaps an unfortunate way of expressing the absolutely correct need for different clinicians to have screens presented that are relevant to the patient and the immediate clinical task at hand. Different clinicians have different needs.
However these are all essentially HUMAN PROBLEMS! Issues about communications, synonyms, value sets, screen display/layout. IT will not solve these issues - as always, the clinicians need to work out the issues amongst themselves. So where CAN we achieve consensus?
Within the openEHR Clinical Knowledge Manager environment we are gaining some traction in achieving clinician agreement to the structure required to define clinical concepts as archetypes – the ‘first principles’ of clinical concepts, if you like. The approach is inclusive of everyone's needs and requirements, rather than requiring an arbitrary decision on the minimum data set or a priority data set - so we aim, as best we can, for a maximum data set.
For example, the framework to express a Diagnosis is largely not contentious:
- Diagnosis name
- Status
- Date of initial onset
- Age at initial onset
- Severity
- Clinical description
- Anatomical location
- Aetiology
- Occurrences
- Exacerbations
- Related problems
- Date of Resolution
- Age at resolution
- Diagnostic criteria
And for a Symptom:
- Symptom name
- Description
- Character
- Duration
- Variation
- Severity
- Current intensity
- Precipitating factors
- Modifying factors
- Course
- Aetiology
- Occurrences
- Previous episodes
- Associated symptoms
This notion of achieving clinician (and other domain expert) consensus and standardisation of clinical concepts is a major focus of the openEHR archetype work.
Bear in mind that while agreement can be achieved on the clinical content structure, this is only the first step in ensuring that clinicians are able to enter, retrieve and exchange meaningful clinical data.
So if we can achieve consensus around these archetypes, do clinicians then have to agree on a standard Discharge Summary or Antenatal Record or Report? The answer: only if is useful to do so. Clinical diversity can be allowed if the archetype pool is stable & governed/managed. By tightly governing the archetypes at international or national level, these are effectively the common 'lingua franca' that enables sharing of health information.
Template creation is the next openEHR layer - these aggregate the archetypes together to represent the requirements for a specific clinical activity and then constrain them down from their fully inclusive state to something that is 'fit for use' for a given clinician in a given clinical situation. Terminology subsets are also integrated in appropriate places into the archetypes (sometimes) and templates (usually) to round out the expressiveness needed by computable clinical models in clinical care - neither the structure nor the terminology can do this in isolation.
Once a critical mass of these archetypes are published we will be able to support a breadth clinical diversity across eHealth projects - the need for rigid, inflexible messages and documents to support any health information exchange has been largely overcome. Clinicians will only need to agree these types of messages or documents where there is a measurable benefit for doing so, and at all other times they can focus on ensuring that they express their archetype-based clinical records as flexibly as they need to for patient care. Sure, the human factors remain unresolved - but not even the most perfect EHR will solve these issues!
So can clinicians agree? Yes! It is happening in many areas related to data structure, creating a solid framework for our electronic health records. Archetypes are the tightly managed building blocks; templates enable safe and flexible expression of clinical and patient records - allowing the best of both worlds, both governance AND clinical diversity to flourish... and the clinicians are finally able to actively participate in shaping their EHRs.
Clinician-led eHealth records - a knowledge-enabled approach
My presentation given to W.H.O. in Geneva last week... [slideshare id=5492832&doc=20101008whoclinician-ledehealthrecords-101019133425-phpapp02]